What is liquid biopsy?
Liquid biopsies enable analysis of biofluids, typically blood, to examine biomarkers shed by solid tumors into circulation1. They can detect actionable genomic alterations in a non-invasive way, providing valuable insights to facilitate early cancer detection and disease monitoring2.
Tumor-derived biomarkers that are a source for liquid biopsy analysis include (Fig. 1):
Circulating tumor cells (CTCs)
CTCs are initially released from primary tumors in the tissue, travel through the bloodstream, and account for the development of metastatic tumors at distant sites in the body. As they are live cells, they have the potential to be used for functional analysis such as therapy sensitivity/resistance evaluation3. However, CTCs are rare events in the blood, which makes them difficult to identify and characterize in routine clinical practice1.
Extracellular vesicles (EV, i.e. microvesicles and exosomes)
EVs are membrane-enclosed structures containing proteins, genetic material, and lipids that can provide biological information on the cell of origin4. Due to their role in pathological processes, EVs are an attractive analyte for liquid biopsy, but their isolation and purification is technically challenging1.
Cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA)
cfDNA refers to DNA fragments that are freely circulating in the bloodstream, primarily originating from normal cells5. Circulating tumor DNA (ctDNA) is the small portion of cfDNA that derives from tumor cells or CTCs undergoing cell death (i.e. apoptosis or necrosis)5. There are well-established methods for isolating cfDNA, and for analyzing it using methods such as PCR and next-generation sequencing (NGS)-based tests6, making it an ideal and feasible substrate for routine genomic analysis.
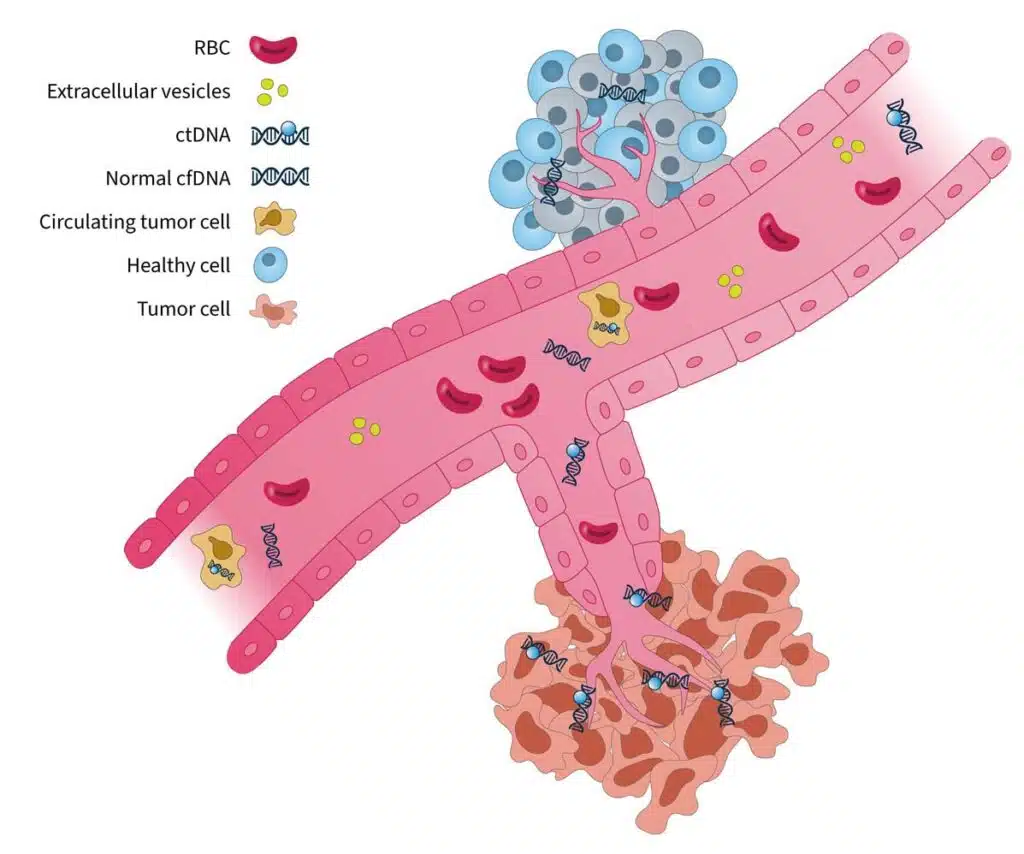
Figure 1. Blood-based cancer biomarkers in liquid biopsy7. RBC, red blood cell.
The analysis of cell-free DNA is a promising method for guiding clinical decisions and can complement current standard-of-care practices8.
What are the clinical applications of liquid biopsy?
In the era of precision medicine, tumor molecular profiling is a critical tool to identify targetable alterations and guide treatment decision-making9. Tissue biopsy is currently the gold standard for tumor profiling8; however, there are limitations associated with this approach:
- Surgery or biopsy to obtain tissue samples can be invasive, expensive, and time-consuming, making it unsuitable for repeat sampling and longitudinal disease monitoring10.
- In certain cancer types, it can be difficult to obtain sufficient tissue quantity and quality10. For example, clinical studies show that up to 20% of non-small cell lung cancers (NSCLC) biopsies are inadequate for molecular analysis11,12.
- The information provided by tissue biopsy is limited to a single point in space and time, failing to represent complex inter- and intra-tumor heterogeneity13.
Liquid biopsy has the potential to be a transformative tool in clinical oncology, showing promise for applications in many stages of cancer management (Fig. 2):
- Liquid biopsy is non-invasive, easily repeatable, and relatively fast to perform8. This makes it an ideal approach for when tissue biopsy is not feasible, or a time-sensitive decision needs to be made.
- It allows real-time monitoring of changes during the course of disease, providing insights into intra-patient spatial and temporal tumor heterogeneity8,13.
- A major advantage of liquid biopsy is its ability to detect minimal residual disease (MRD)14, which is not detectable by conventional routine tests. MRD refers to the small number of cancer cells that remain in the body following treatment that can drive the reemergence of cancer14. Measurement of MRD using liquid biopsy can provide valuable insights into treatment response and risk of cancer recurrence14.
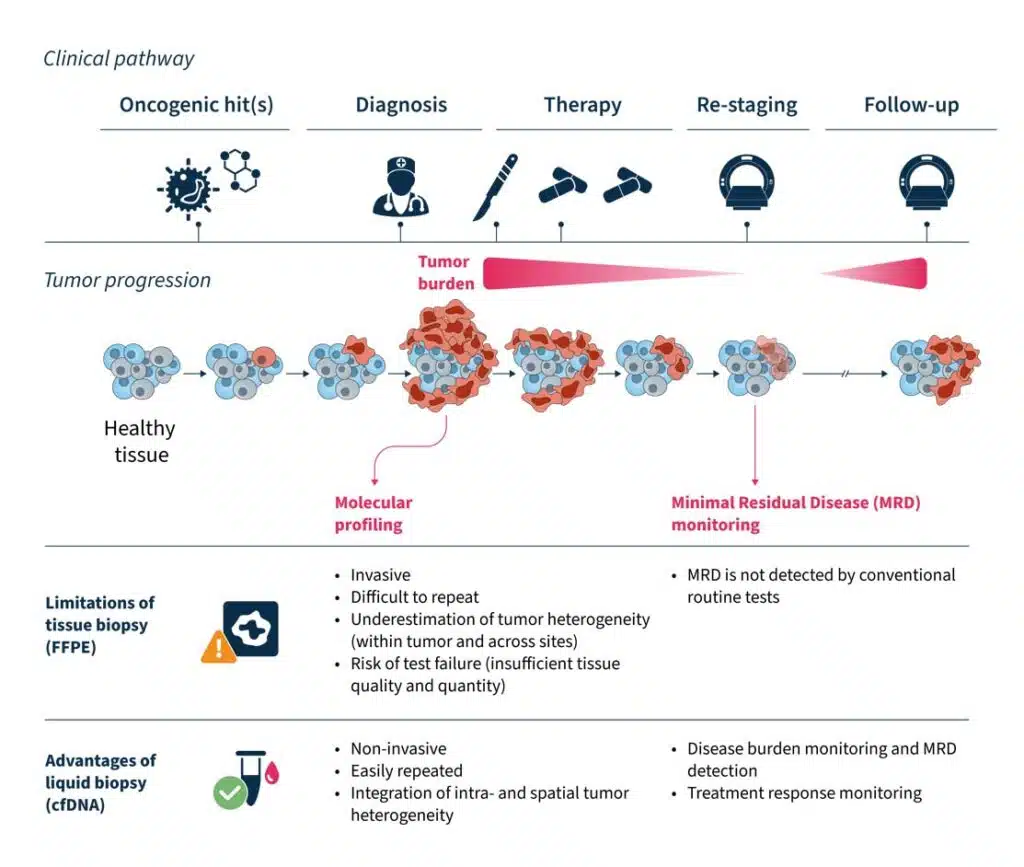
Figure 2. The advantages and clinical utility of liquid biopsy in the cancer care journey10–15.
Innovations in liquid biopsy analysis over the past decade have led to the regulatory approvals of blood-based tests to guide treatment for NSCLC, prostate, breast, and ovarian cancers16. Clinical guidelines have also provided expert recommendations for its use in specific clinical scenarios8,15. Despite great advances in technology and its increasing utility in clinical practice, there are still challenges to overcome when using liquid biopsy to identify clinically relevant information.
Overcoming “fisherman’s luck” in liquid biopsy
One challenging aspect of liquid biopsy analysis is that ctDNA concentration varies greatly across cancer types and between patients17. In patients with cancer, the quantity of ctDNA in the blood can be impacted by several factors, including histology, tumor site, clinical factors (age, sex, treatment history, etc.), and ctDNA fragmentation17. Therefore, it is important to have a robust test to detect clinically relevant variants, even at low ctDNA concentrations against a cfDNA background.
Another factor that may impact liquid biopsy analysis is the presence of clonal hematopoiesis of indeterminate potential (CHIP). In healthy individuals, the majority of cfDNA arises from hematopoietic cells (i.e. stem cells in the bone marrow that give rise to other blood cells)18. Normal hematopoietic cells accumulate somatic mutations during aging, known as CHIP, which are technically indistinguishable to tumor-specific mutations in NGS assays18,19. It is important that the biological noise caused by CHIP signals are removed in liquid biopsy analysis to eliminate false positive variant calls and give an accurate representation of disease burden19,20.
These biological confounders can make “fishing” for clinically relevant information in cfDNA a challenge (Fig. 3). For example, if a patient has high disease burden, there is likely more ctDNA available to analyze, which makes it easier to “catch” what you are looking for. However, if there is less ctDNA and more biological noise, you may need to modify your tools and approach to improve your yield.
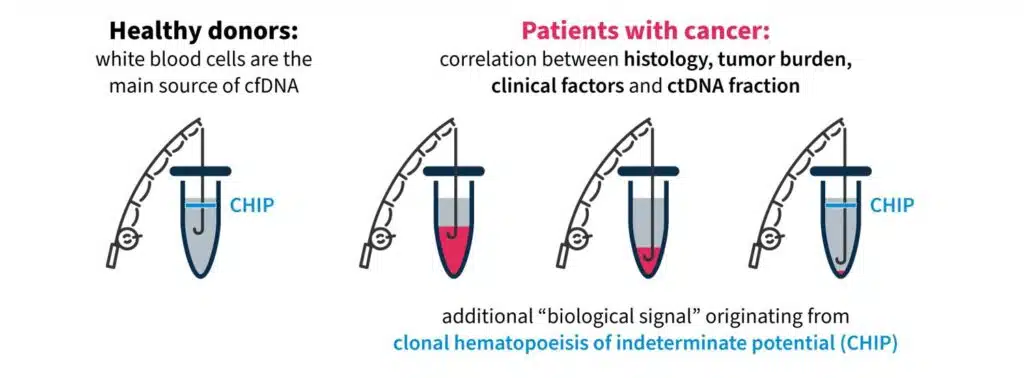
Figure 3. “Fishing” for clinically relevant information in liquid biopsy can be complicated by biological confounders17,18.
Highly precise and sensitive liquid biopsy technologies are needed to overcome “fisherman’s luck” and detect rare, causative variants and disease burden in cfDNA. Guidelines issued by the ESMO Precision Medicine Working Group on the use of cfDNA assays in clinical practice discuss the need for advanced techniques capable of capturing spatial and temporal tumor heterogeneity and reducing rates of false negatives8.
Pioneer innovation with SOPHiA DDMTM for Liquid Biopsy
SOPHiA GENETICS is at the forefront of innovation in liquid biopsy technology for tumor profiling. The advanced proprietary algorithms of the SOPHiA DDMTM Platform empower clinical researchers to reveal deep genomic insights from cell-free DNA samples.
With a streamlined, sample-to-report NGS workflow, you can:
- Retain custody of your cell-free DNA samples and your data: Analyze your samples directly in your lab, combining an in-house wet lab workflow with the cloud-based analytics of the SOPHiA DDM™ Platform.
- Customize your application to focus on the biomarkers that matter most to you: Develop a high-quality application that suits your needs and receive comprehensive support at every step with the SOPHiA DDM™ MaxCare Program.
- Reduce noise with our proprietary molecular barcoding technology: Detect variants at low frequency (0.5% VAF) starting from 25 ng cell-free DNA with our proprietary unique molecular identifier (UMI) technology, CUMIN™.
In addition, we are excited to be collaborating with Memorial Sloan Kettering Cancer Center (MSK) to decentralize MSK-ACCESS® for liquid biopsy, designed to provide a maximum coverage of cancer disease variants in ctDNA20. By combining MSK’s clinical expertise in cancer genomics, the predictive algorithms of the SOPHiA DDMTM Platform, and the power of the global SOPHiA GENETICS community, the collaboration aims to expand access to precision cancer analysis capabilities worldwide.
Read more about how you can enhance your analytical capabilities and advance your clinical research here.
References
- Marrugo-Ramírez J, Mir M, Samitier J. Int J Mol Sci. 2018;19(10):2877.
- Ou SI, Nagasaka M, Zhu VW. Am Soc Clin Oncol Educ Book. 2018;38:978-997.
- Eslami-S Z, Cortés-Hernández LE, Thomas F, et al. Br J Cancer. 2022;127(5):800-810.
- Liu J, Chen Y, Pei F, et al. Biomed Res Int. 2021;2021:6611244.
- Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Cancer Biol Ther. 2019;20(8):1057-1067.
- Chen M, Zhao H. Hum Genomics. 2019;13(1):34.
- Adapted from ‘CtDNA in circulation’, made available by Wikimedia Commons under the Creative Commons Attribution-Share Alike 4.0 International license.
- Pascual J, Attard G, Bidard FC, et al. Ann Oncol. 2022;33(8):750-768.
- Mosele F, Remon J, Mateo J, et al. Ann Oncol. 2020;31(11):1491-1505.
- Di Sario G, Rossella V, Famulari ES, et al. Front Genet. 2023;14:1152470.
- Arcila ME, Oxnard GR, Nafa K, et al. Clin Cancer Res. 2011;17(5):1169-1180.
- Sholl LM, Aisner DL, Varella-Garcia M, et al. J Thorac Oncol. 2015;10:768–77.
- Russano M, Napolitano A, Ribelli G, et al. J Exp Clin Cancer Res. 2020;39(1):120
- Pantel K, Alix-Panabières C. Nat Rev Clin Oncol. 2019;16(7):409-424.
- García-Pardo M, Makarem M, Li JJN, Kelly D, Leighl NB. Br J Cancer. 2022;127(4):592-602.
- Heitzer E, van den Broek D, Denis MG, et al. ESMO Open. 2022;7(2):100399.
- Hsiehchen D, Espinoza M, Gerber DE, Beg MS. Cancer Biol Ther. 2021;22(7-9):455-464.
- Abbosh C, et al. Ann Oncol. 201; 30(3): 358–359.
- Razavi P, et al. Nat Med. 2019 Dec;25(12):1928-1937.
- Brannon AR, et al. Nat Commun. 2021;18;12(1):3770.