Deeper genomic insights, fewer missed opportunities
CGP enables clinical researchers to identify actionable variants and biomarkers across hundreds of genes using a single solution. The SOPHiA DDMTM Platform offers decentralized, in-house next generation sequencing (NGS) applications that help maximize insights from CGP data by leveraging advanced analytical capabilities, intutive interpretation features, and one-click reporting.
Benefits of SOPHiA DDM™ for CGP
Retain ownership of your samples and data by bringing CGP workflows in-house.
Stay ahead with globally curated gene content
To facilitate variant annotation and interpretation, CGP is increasingly being included in guidelines and recommendations1,2. Our CGP applications are designed to cover genes of emerging and known associations to multiple tumor types, ensuring that you stay ahead of the changing guidelines landscape in your cancer research.
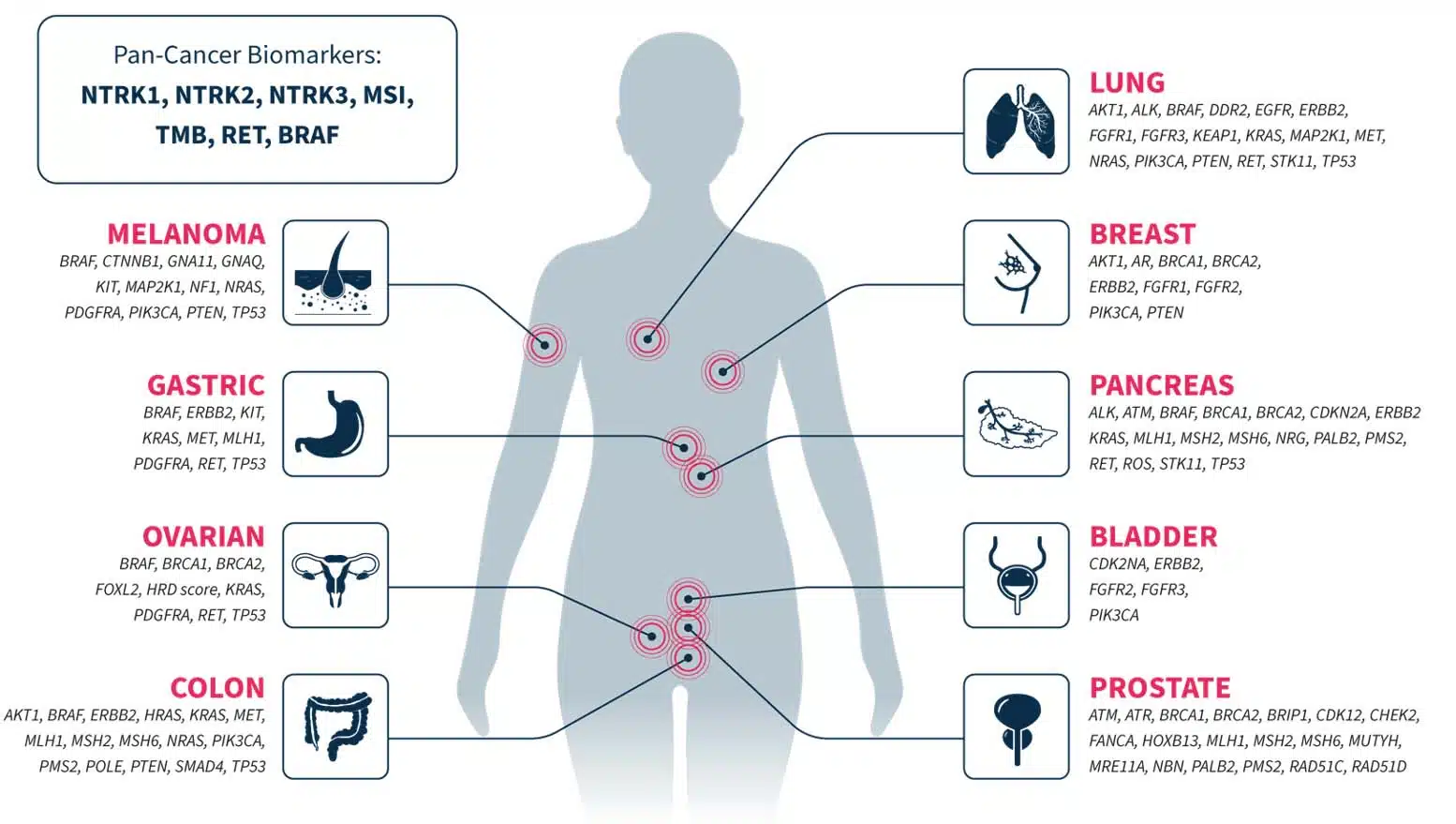
Not an exhaustive list of genes or biomarkers.
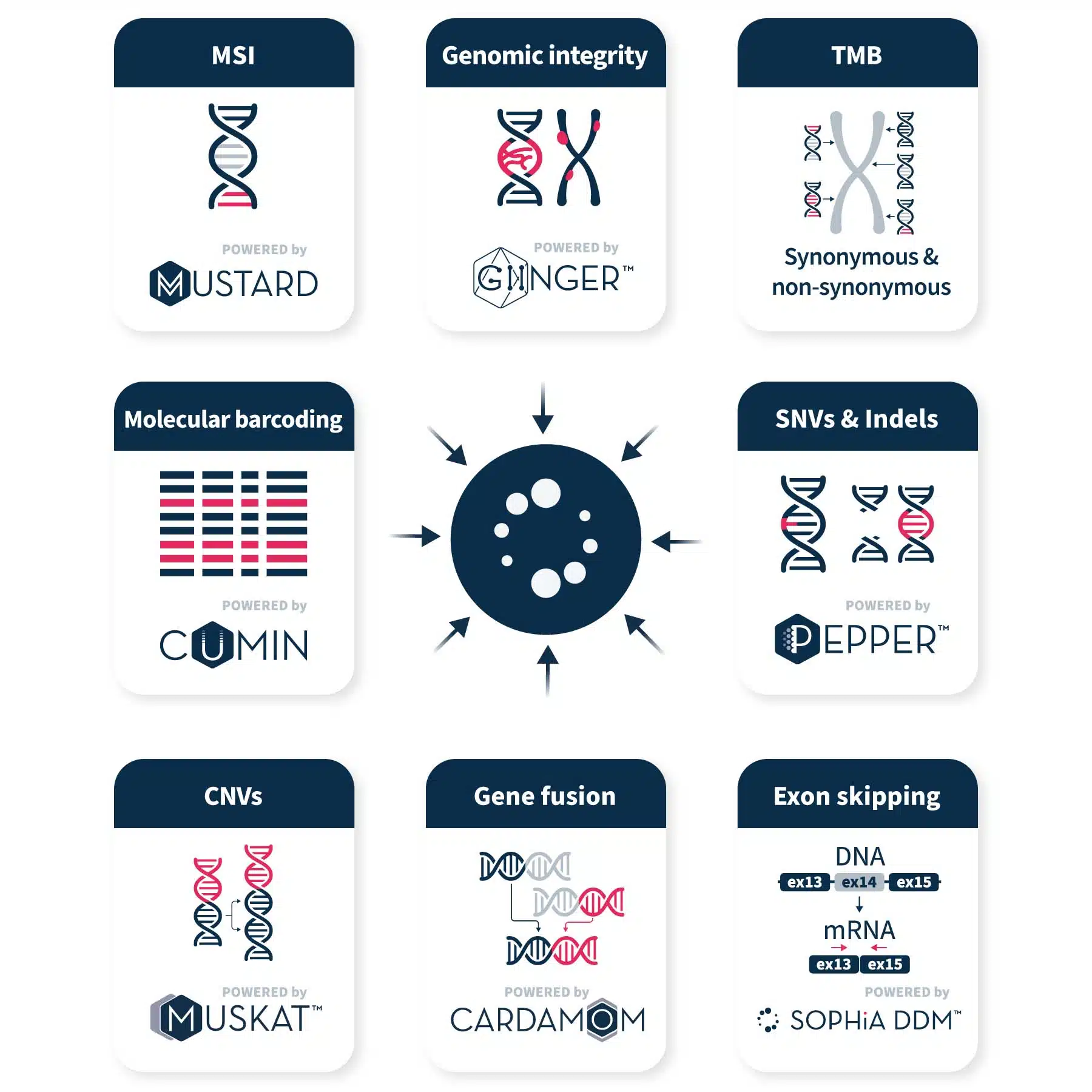
Uncover insights with advanced analytical technology
The SOPHiA DDM™ Platform uses proprietary algorithms to efficiently call, annotate, and pre-classify variants and biomarkers, including single nucleotide variants (SNVs), copy number variations (CNVs), insertions and deletions (Indels), gene fusions, microsatellite instability (MSI), genomic integrity and tumor mutational burden (TMB) from raw NGS data.
Our CGP applications supports accurate variant calling and biomarker detection in key genes associated with solid tumors, allowing you to pinpoint the most relevant alterations for your lab.
SOPHiA DDM™ demonstrated high correlation of overall (R2 = 0.99) and non-synonymous (R2 = 0.99) TMB estimation when compared to established reference method4.
Based on concordance with TSO500 LocalApp v2.2 comparing Mut/Mb in 64 samples, including 53 clinical FFPE tissue samples from more than 10 tumor types.

SOPHiA DDM™ RNAtarget Technology demonstrated high analytical sensitivity (100%) and specificity (99.99%) for novel (partner-agnostic) fusion detection in RNA and tNA samples5.

Focus on what matters with evidence-based decision support
OncoPortal™ Plus matches tumor molecular profiles with
clinical associations and available clinical trials,
leveraging expertly curated evidence powered by JAX-CKB™.
After interpretation, the flexible reporting tools enable
users to prepare one-click, comprehensive reports that save time and are customizable to your needs.

Save time with a streamlined workflow
The SOPHiA DDM™ Platform offers in-house workflows that are expertly designed to facilitate the analysis and interpretation of relevant variants and biomarkers from DNA and RNA. With both bundle and dry lab applications available, we provide CGP options that suit your lab’s unique set-up and requirements.

Adoption of our genomic applications is made easy with the SOPHiA DDM™ MaxCare Program, empowering teams to gain confidence in their results, establish workflow efficiency, meet quality standards, and ensure performance by offering comprehensive training and analytical performance assessment.
Interested in complementing your CGP workflow with liquid biopsy analysis? Advance your oncology research to new horizons with SOPHiA DDM™ for Liquid Biopsy.
Specifications
Bundle solution
(sample-to-report)
Bundle solution
(sample-to-report)
Dry lab solution
(FASTQ-to-report)
Dry lab solution
(FASTQ-to-report)” row_type=”head” _builder_version=”4.23.1″ _module_preset=”default” global_colors_info=”{}”][/difl_datatableitem][difl_datatableitem row=”
MSK-IMPACT® powered with SOPHiA DDM™
In developmentᵃ
SOPHiA DDM™ Cancer Profiling Solution
SOPHiA DDM™ for TSO500
SOPHiA DDM™ for SureSelect Cancer CGP Assay” _builder_version=”4.24.0″ _module_preset=”default” row_text_font=”|700|||||||” global_colors_info=”{}”][/difl_datatableitem][difl_datatableitem row=”No.of genes
505 DNA
• 480 DNA
• 136 RNA
• 556 total unique genes
• 523 DNA
• 55 RNA
• 679 DNA
• 80 RNA” _builder_version=”4.24.0″ _module_preset=”default” global_colors_info=”{}”][/difl_datatableitem][difl_datatableitem row=”Diseases Covered
Multi-cancer (any solid tumor)
Multi-cancer (any solid tumor)
Multi-cancer (any solid tumor)
Multi-cancer (any solid tumor)” _builder_version=”4.23.1″ _module_preset=”default” global_colors_info=”{}”][/difl_datatableitem][difl_datatableitem row=”Sample Type
FFPE and matched normal blood sample
FFPE
FFPE
Fresh-frozen, FFPE” _builder_version=”4.23.1″ _module_preset=”default” global_colors_info=”{}”][/difl_datatableitem][difl_datatableitem row=”Recommended Input Amount
TBD
50 ng DNA, 50 ng RNA
40ng DNA, 40ng RNA
50 ng DNA, 50 ng RNA” _builder_version=”4.24.2″ _module_preset=”default” hover_enabled=”0″ global_colors_info=”{}” sticky_enabled=”0″][/difl_datatableitem][difl_datatableitem row=”Sequencer Compatibility
• Illumina® NextSeq® 550/1000/2000
• Illumina® NovaSeq™
• Illumina® NextSeq® 550/1000/2000
• Illumina® NovaSeq™
• Illumina® NextSeq® 550/1000/2000
• Illumina® NovaSeq™
• Illumina® NextSeq® 550/1000/2000
• Illumina® NovaSeq™” _builder_version=”4.24.2″ _module_preset=”default” hover_enabled=”0″ global_colors_info=”{}” sticky_enabled=”0″][/difl_datatableitem][difl_datatableitem row=”Detected Variants & Biomarkers
• SNVs and Indels
• CNVs
• Whole gene amplifications and deletions
• DNA translocations/gene fusions
• TMB
• MSI
• SNVs and Indels
• CNVs (454 genes)
• Whole gene amplifications and deletions
• Novel fusions/splice variants (136 genes)
• TMB
• MSI
• SNVs and Indels
• CNVs/amplifications (495 genes)
• Novel fusions/splice variants (55 genes)
• TMB
• MSI
• SNVs and Indels
• CNVs/amplications (32 genes)
• Translocations (12 genes)
• Novel fusions/splice variants (80 genes)
• TMB
• MSI” _builder_version=”4.24.2″ _module_preset=”default” hover_enabled=”0″ global_colors_info=”{}” sticky_enabled=”0″][/difl_datatableitem][/difl_datatable]
Bundle solution (sample-to-report)
MSK-IMPACT® powered with SOPHiA DDM™ ᵃ
[difl_datatable body_cell_padding="20px||20px||true|false" module_class="liquidtable" _builder_version="4.23.1" _module_preset="default" head_text_font="|700|||||||" head_text_text_align="center" head_text_text_color="#0A2E4A" head_text_font_size="18px" row_text_font="|300|||||||" row_text_text_align="center" row_text_font_size="18px" row_first_column_text_font="|700|||||||" row_first_column_text_text_align="left" background_color="#FFFFFF" hover_enabled="0" row_text_text_align_tablet="center" row_text_text_align_phone="left" row_text_text_align_last_edited="on|phone" template_type="module" global_colors_info="{}" sticky_enabled="0"][difl_datatableitem enable_column_merge="on" body_col_span_item="4" body_col_span_item_value="2" row="
" row_type="head" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="No. of genes
505 DNA" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Diseases Covered
Multi-cancer (any solid tumor)" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Sample Type
FFPE and matched normal blood sample" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Recommended Input Amount
TBD" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Sequencer Compatibility
• Illumina NextSeq® 550/1000/2000
• Illumina® NovaSeq" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Detected Variants & Biomarkers
• SNVs and Indels
• CNVs
• DNA translocations/gene fusions
• TMB
• MSI" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][/difl_datatable]
SOPHiA DDM™ Cancer Profiling Solution
[difl_datatable body_cell_padding="20px||20px||true|false" module_class="liquidtable" _builder_version="4.23.1" _module_preset="default" head_text_font="|700|||||||" head_text_text_align="center" head_text_text_color="#0A2E4A" head_text_font_size="18px" row_text_font="|300|||||||" row_text_text_align="center" row_text_font_size="18px" row_first_column_text_font="|700|||||||" row_first_column_text_text_align="left" background_color="#FFFFFF" row_text_text_align_tablet="center" row_text_text_align_phone="left" row_text_text_align_last_edited="on|phone" template_type="module" global_colors_info="{}" theme_builder_area="post_content"][difl_datatableitem enable_column_merge="on" body_col_span_item="4" body_col_span_item_value="2" row="
" row_type="head" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem row="No. of genes
• 480 DNA
• 136 RNA" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem row="Diseases Covered
Multi-cancer (any solid tumor)" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem row="Sample Type
FFPE" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem row="Recommended Input Amount
50 ng DNA, 50 ng RNA" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Sequencer Compatibility
• Illumina® NextSeq® 550
• Illumina® NovaSeq" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Detected Variants & Biomarkers
• SNVs and Indels
• CNVs (454 genes)
• Whole gene amplifications and deletions
• Novel fusions/splice variants
• TMB
• MSI" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}" theme_builder_area="post_content"][/difl_datatableitem][/difl_datatable]
Dry lab solution (FASTQ-to-report)
SOPHiA DDM™ for TSO500
[difl_datatable body_cell_padding="20px||20px||true|false" module_class="liquidtable" _builder_version="4.23.1" _module_preset="default" head_text_font="|700|||||||" head_text_text_align="center" head_text_text_color="#0A2E4A" head_text_font_size="18px" row_text_font="|300|||||||" row_text_text_align="center" row_text_font_size="18px" row_first_column_text_font="|700|||||||" row_first_column_text_text_align="left" background_color="#FFFFFF" hover_enabled="0" row_text_text_align_tablet="center" row_text_text_align_phone="left" row_text_text_align_last_edited="on|phone" template_type="module" global_colors_info="{}" sticky_enabled="0"][difl_datatableitem enable_column_merge="on" body_col_span_item="4" body_col_span_item_value="2" row="
" row_type="head" _builder_version="4.23.1" _module_preset="default" global_colors_info="{}"][/difl_datatableitem][difl_datatableitem row="No. of genes
• 523 DNA
• 55 RNA" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Diseases Covered
Multi-cancer (any solid tumor)" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Sample Type
FFPE" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Recommended Input Amount
40ng DNA, 40ng RNA" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Sequencer Compatibility
• Illumina NextSeq® 550/1000/2000
• Illumina® NovaSeq" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Detected Variants & Biomarkers
• SNVs and Indels
• CNVs/amplifications (495 genes)
• Novel fusions/splice variants (55 genes)
• TMB
• MSI" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][/difl_datatable]
SOPHiA DDM™ for SureSelect Cancer CGP Assay
[difl_datatable body_cell_padding="20px||20px||true|false" module_class="liquidtable" _builder_version="4.23.1" _module_preset="default" head_text_font="|700|||||||" head_text_text_align="center" head_text_text_color="#0A2E4A" head_text_font_size="18px" row_text_font="|300|||||||" row_text_text_align="center" row_text_font_size="18px" row_first_column_text_font="|700|||||||" row_first_column_text_text_align="left" background_color="#FFFFFF" hover_enabled="0" row_text_text_align_tablet="center" row_text_text_align_phone="left" row_text_text_align_last_edited="on|phone" template_type="module" global_colors_info="{}" sticky_enabled="0"][difl_datatableitem enable_column_merge="on" body_col_span_item="4" body_col_span_item_value="2" row="
" row_type="head" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="No. of genes
• 679 DNA
• 80 RNA" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Diseases Covered
Multi-cancer (any solid tumor)" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Sample Type
Fresh-frozen, FFPE" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem row="Recommended Input Amount
50 ng DNA, 50 ng RNA" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Sequencer Compatibility
• Illumina® NextSeq® 550/1000/2000
• Illumina® NovaSeq" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][difl_datatableitem enable_column_merge="on" body_col_span_item="3" body_col_span_item_value="3" row="Detected Variants & Biomarkers
• SNVs and Indels
• CNVs/amplifications (32 genes)
• Translocations (12 genes)
• Novel fusions/splice variants (80 genes)
• TMB
• MSI" _builder_version="4.23.1" _module_preset="default" hover_enabled="0" global_colors_info="{}" sticky_enabled="0"][/difl_datatableitem][/difl_datatable]
ᵃProduct in development – Technology and concepts in development. May not be available for sale.
Resources
Get in touch to find out more
Our client services team is on hand to answer any questions or schedule your live demo.
References
- Mosele F, et al. Ann Oncol. 2020;31(11):1491–505.
- Chakravarty D, et al. J Clin Oncol. 2022;40(11):1231–58.
- Marques A, et al. Front Oncol. 2022;12:9692.
- Data on file.
- Data on file.

