SOPHiA DDM™ Whole Exome Solution v2
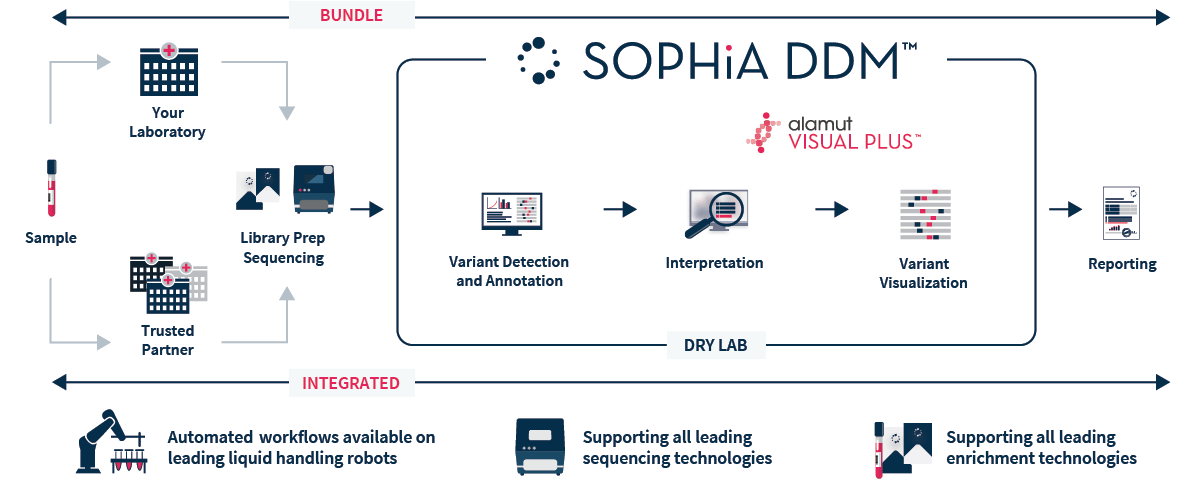
SOPHiA DDM™ Whole Exome Solution v2 is an NGS-based application that provides a streamlined end-to-end workflow, from sample to variant report, to accelerate rare and inherited disease research.
SOPHiA DDM™ Whole Exome Solution v2 targets nuclear genes and the full mitochondrial genome to offer a single expertly designed capture-based target enrichment kit. This updated gene content is combined with the analysis and interpretation capabilities of the SOPHiA DDM™ platform, offering uniform coverage of the targeted regions within a streamlined workflow. Our cloud-based platform helps you greatly increase the efficiency of your laboratory by reducing the burden of data interpretation and storage, thus optimizing time and resources.
Expertly designed panel with 19,425 genes optimized to provide uniform coverage across GC-rich regions for confident variant detection, including CNV detection in 97% of genes
Full mitochondrial genome coverage, including heteroplasmy analysis, for increased sensitivity and precision for complex, heterogeneous variants
Ready-to-sequence target-enriched library in 8 hours hands-on time
Accurate variant annotation based on UCSC-built hg38 human genome and comprehensive transcript annotation with MANE
Streamlined variant interpretation with the SOPHiA DDM™ platform’s integrated features, including trio analyses to analyze variants by inheritance mode and access to the most updated and trusted databases, such as OMIM® and HPO
Secure and scalable cloud-based data storage with secure knowledge sharing
Product Details
The coverage you need for accurate CNV detection
SOPHiA DDM™ Whole Exome Solution v2 achieves high on-target rates that ensure reliable coverage uniformity values across all the target regions, even in GC-rich regions. Equal read coverage is crucial for the precise identification of multiple types of variations, including copy number variations (CNVs). With SOPHiA DDM™ Whole Exome Solution v2, you can reach more than 99% analytical sensitivity* for CNV detection.
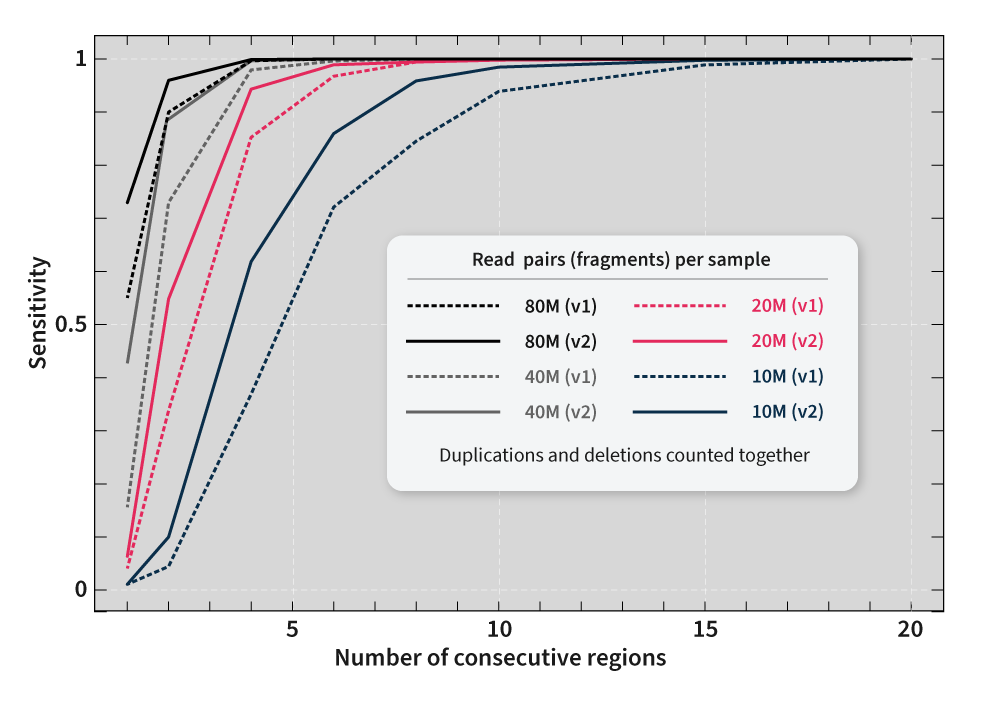
*Analytical performance for CNVs was calculated with 80M reads per sample. Sequencing was performed using an Illumina NovaSeq® instrument.
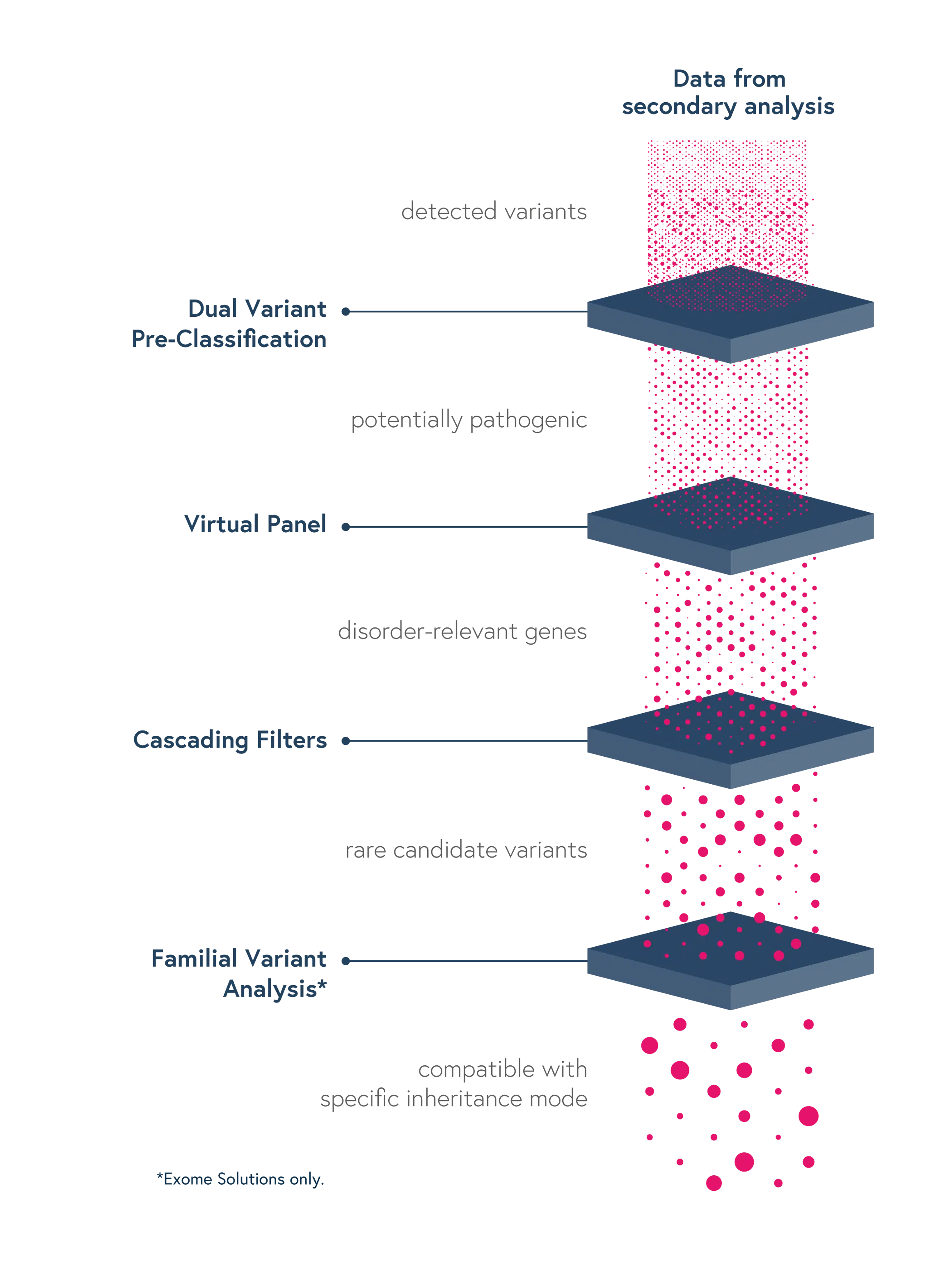
Dedicated features to ease variant interpretation
Dedicated features in SOPHiA DDM™ reduce the complexity of determining the significance of genomic variants and facilitate the interpretation process, thus reducing turnaround time:
- GRCh38/hg38 based analytics – Annotate variants accurately
- Dual Variant Pre-classification – Improve assessment of variant pathogenicity with both ACMG scores and SOPHiA GENETICS machine learning-based predictions
- Virtual Panels – Restrict the interpretation to sub-panels of genes of interest using the HPO or OMIM® browser
- Cascading Filters – Apply custom filtering options for quicker screening of relevant variants and save strategies for future analyses
- Familial Variant Analysis (trio-analysis) – Identify pathogenic variants considering different modes of inheritance, through a family-based approach
Through SOPHiA DDM™, you can also access Alamut™ Visual Plus, a full-genome browser that integrates numerous curated genomic and literature databases, guidelines, missense and slicing predictors, thus enabling a deeper variant exploration.
Specifications
| Parameters | Details |
|---|---|
| Genes | 19,425 |
| Sample Type | Blood |
| DNA Input Amount | 200 ng |
| Sequencer Compatibility |
|
| Library Preparation Time | 8 hours Hands-on time |
| Analysis Time From FASTQ File | Overnight |
| Detected Variants |
|
Resources
Data sheets
Fact sheets
Related webinars
Contact us
Please fill out the form below to get in touch
Related products
SOPHiA DDM™️ Clinical Exome Solution v3
Facilitate the assessment of challenging cases of Mendelian diseases with our end-to-end workflow that reduces costs and allows faster completion of studies.

SOPHiA DDM™ for KAPA HyperExome
With a fully integrated workflow from FASTQ to report, our solution streamlines multiple types of variants detection, interpretation, and reporting.
