SOPHiA DDM™ Clinical Exome Solution v3
With new and improved probe design, rare and inherited disorder analyses benefit from increased detection capabilities within the entire mitochondrial genome, as well as non-coding variants known to be disorder-associated, for a deeper investigation of Mendelian disorders.
SOPHiA DDM™ Clinical Exome Solution v3 offers a streamlined end-to-end workflow (from sample to variant report), that streamlines and facilitates the assessment of challenging Mendelian disorders, freeing up time and resources. The solution bundles a capture-based target enrichment kit with the analytical capabilities and interpretation-support functionalities of SOPHiA DDM™ platform, offering deep coverage of the target regions and accurate analysis of multiple type of variants (SNVs, Indels and CNVs) in one unique experiment. As a result, we help you efficiently find the pathogenic needle in the big data haystack, and improving turnaround time.
Accurate variant calling including CNV detection and annotation powered by GRCh38/hg38 based analytics
Streamlined variant interpretation thanks to the SOPHiA DDM™ platform-integrated prioritization and filtering features, including trio analyses to analyze variants by inheritance mode, and access to updated and trusted databases, such as OMIM® and HPO
Ready-to-sequence target enriched library in 8 hours hands-on time
Expertly designed panel with 5,500 genes, including the entire mitochondrial genome and ~ 200 non-coding variants with known pathogenicity in deep introns/enhancer/promoter genes
Product Details
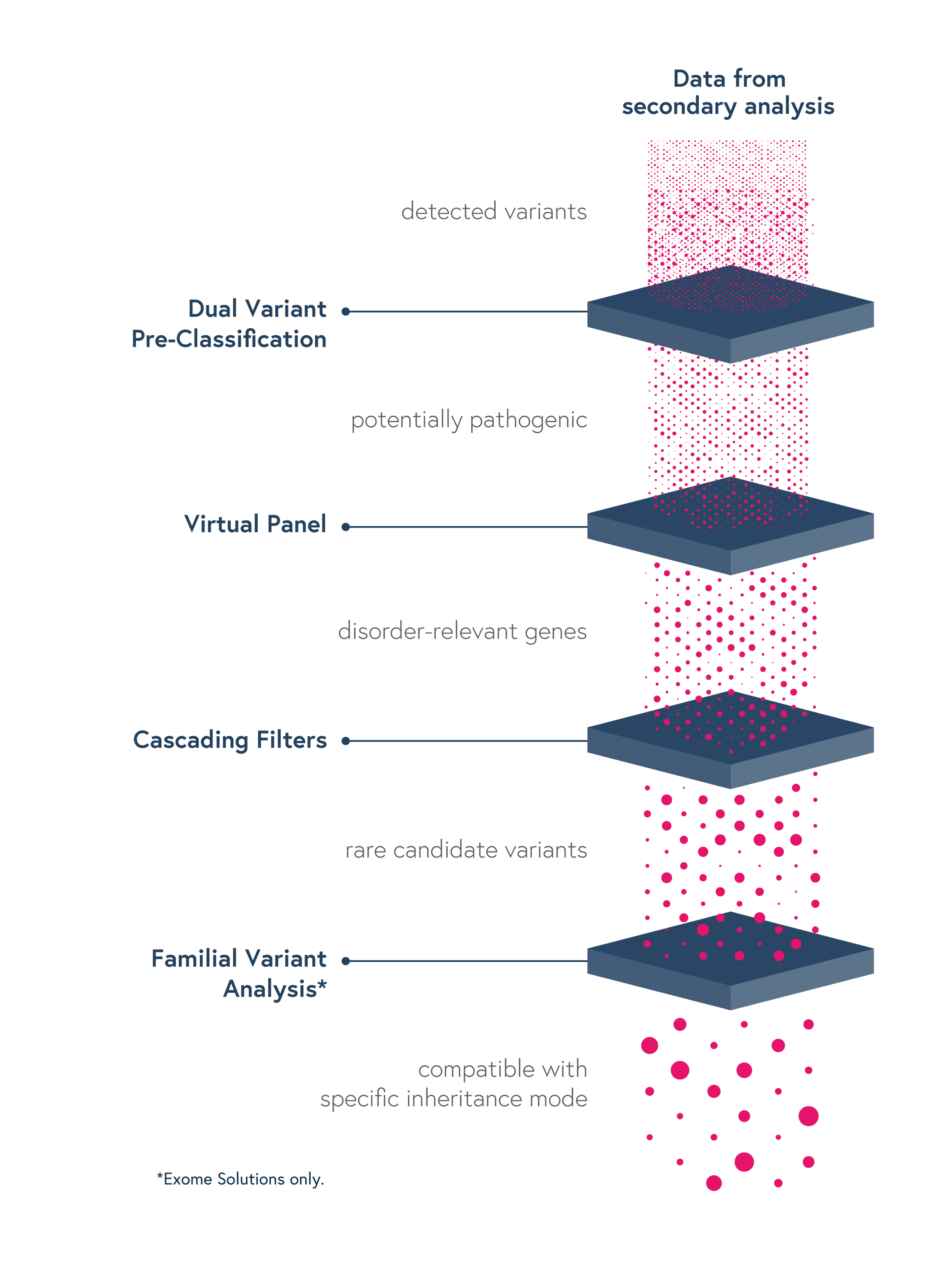
Dedicated features to ease variant interpretation
Dedicated features in SOPHiA DDM™ reduce the complexity of determining the significance of genomic variants and facilitate the interpretation process, thus reducing turnaround time:
-
GRCh38/hg38 based analytics – Annotate variants accurately
- Dual Variant Pre-classification – Improve assessment of variant pathogenicity with both ACMG scores and SOPHiA GENETICS machine learning-based predictions
- Virtual Panels – Restrict the interpretation to sub-panels of genes of interest using the HPO or OMIM® browser
- Cascading Filters – Apply custom filtering options for quicker screening of relevant variants and save strategies for future analyses
- Familial Variant Analysis (trio-analysis) – Identify pathogenic variants considering different modes of inheritance, through a family-based approach
Through SOPHiA DDM™, you can also have access to Alamut™ Visual Plus, a full-genome browser that integrates numerous curated genomic and literature databases, guidelines, missense and slicing predictors, thus enabling a deeper variant exploration.
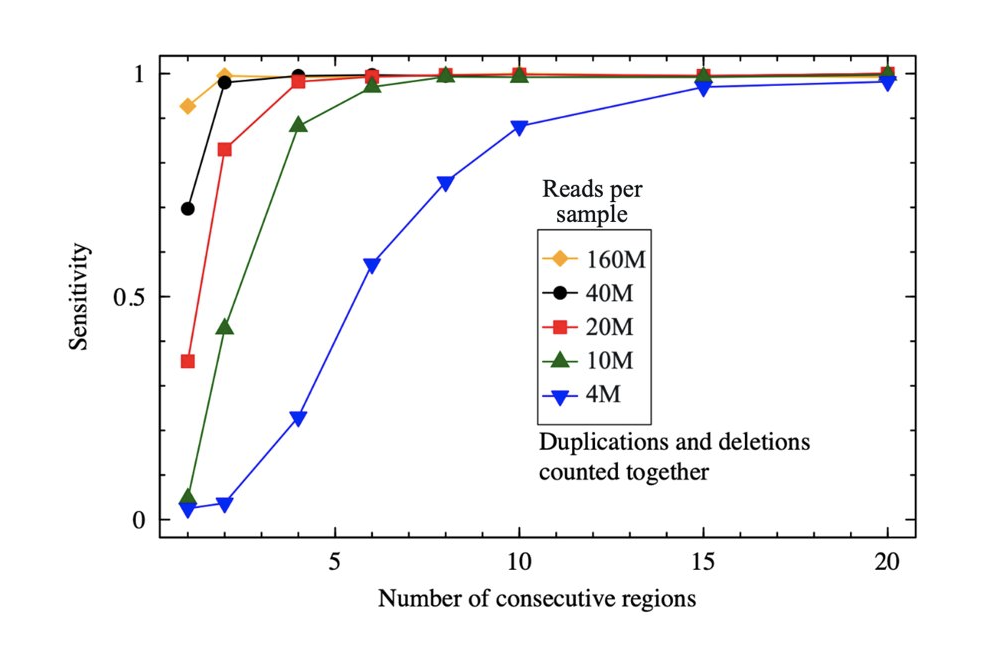
Full coverage of the mitochondrial genome coupled with sensitive variant calling
The probe design is highly optimized to guarantee high on-target reads percentage and coverage uniformity even in GC-rich regions, including the first exon. With SOPHiA DDM™ Clinical Exome Solution v3, you get full mitochondrial genome variant detection, including non-coding variants. Additionally, with improved analytical performance, you can reach 98% analytical sensitivity for CNV detection*.
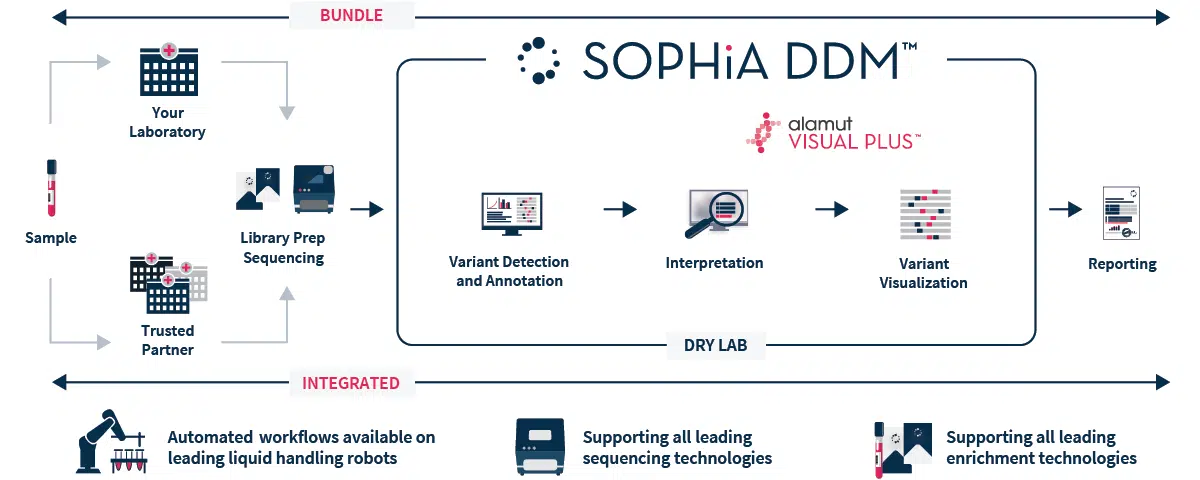
Specifications
| Attribute | Content |
|---|---|
| Addressed Diseases | Rare and inherited disorders |
| Genes | 5,500 Entire mitochondrial genome ~ 200 non-coding variants with known pathogenicity in deep introns/enhancer/promoter genes |
| Target Region Size | 16 Mb |
| Sample Type | Blood |
| DNA Input Amount | 200 ng |
| Sequencer Compatibility |
|
| Library Preparation Time | 8 hours hands-on time |
| Analysis Time From FASTQ File | 6 hours |
| Detected Variants |
|
Resources
Data sheets
Contact us
Please fill out the form below to get in touch
Related products
SOPHiA DDM™ Whole Exome Solution v2
Our sample-to-report solution supports researchers in accelerating assessment of variants associated with rare and inherited diseases.

SOPHiA DDM™ for KAPA HyperExome
With a fully integrated workflow from FASTQ to report, our solution streamlines multiple types of variants detection, interpretation, and reporting.

Footnotes
(*Data on file. Analytical performance for CNVs was calculated on 80 CNVs, sequenced on an Illumina NextSeq® instrument.)