Menu
We recently presented two posters at the European Congress of Pathology 2024, in Florence, Italy, in collaboration with Janssen Pharmaceutical, a Johnson & Johnson company.
The posters provided real-world data insights on the genomic landscape of specific biomarkers associated with lung and prostate cancers, in a subset of European countries.
The first poster spotlight features Stefano Cheloni, Senior Bioinformatician - Tertiary Analysis at SOPHiA GENETICS, presenting the poster titled: “Real-world insights from France, Italy, Spain, and Austria for the investigation of common (exons 19,21) and rare (exons 18, 20) EGFR mutations in lung cancer”.
The identification of exon-specific EGFR mutations can guide the appropriate treatment of lung cancer with tyrosine kinase inhibitors (TKIs) or alternative targeted therapies. This project aimed to explore the landscape of next-generation sequencing (NGS) testing practices for EGFR mutations in clinical practice in a subset of European countries, to determine the potential number of individuals with lung cancer that could benefit from treatment with TKIs or alternative targeted therapies.
Read the EGFR poster through here.
Watch the spotlight below:
In our second poster spotlight, Adrian Janiszewski, Manager Bioinformatician - Team Lead Tertiary Analysis at SOPHiA GENETICS, showcases a poster titled: “NGS testing practices and molecular profiles of BRCA1/2 in prostate cancer: Real-world insights from France, Italy, Spain, and Austria”.
BRCA1/2 testing can inform which prostate cancer patients might respond to PARP inhibitors (PARPi). Having a better understanding of BRCA1/2 testing practices in the real-world setting can provide valuable insights into identifying gaps and opportunities to improve the identification of metastatic prostate cancer (mPC) patients who may benefit from PARPi treatment. In this study we investigated NGS testing practices results across specific European countries.
Read the BRCA 1 & 2 poster through here.
Watch the spotlight below:
We would like to warmly thank Stefano and Adrian for their insightful presentations and for sharing the key takeaways of these projects.
Learn more about SOPHiA DDM™ for BioPharma, by visiting the page.
DISCLAIMER:
All product and company names are trademarks™ or registered trademarks of their respective holders. Use of them does not imply any affiliation with or endorsement by them. SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
Tell us a bit about your laboratory.
The Genetics and Genomics Laboratory in which we operate belongs to a Local Health Authority in the Sardinia region. We provide molecular evaluation of gene alterations in many solid neoplasms to identify patients eligible for personalized treatments (i.e. targeted therapy). This service is provided to medical oncology departments in the city of Cagliari and many peripheral areas in the region.
What are the roots of your collaboration with SOPHiA GENETICS, and which application are you using?
The decision to collaborate with SOPHiA GENETICS was made in 2019, as our request to activate our NGS service was subject to CE-IVD certification of the library preparation and sequencing path. Furthermore, the process seemed quite linear to us.
We are currently using the SOPHiA DDM™ RNAtarget Oncology Solution (ROS) for RNA sequencing.
Can you describe your experience of using SOPHiA DDM™ ROS in routine for your RNA sequencing?
It has an excellent ability to identify fusion gene events involved in many oncological diseases. The application allowed us to identify several known and unknown molecular alterations (single nucleotide variants, Indels, gene fusion and exon skipping events) involved in various cancers, which represent biomarkers for approved and agnostic target drugs. In this way, it is possible to perform a combined analysis of different tumor pathologies in a single analysis session.
“It has an excellent ability to identify fusion gene events involved in many oncological diseases.”
Can you describe your experience with automating the workflow?
The automation of the workflow for library preparation has allowed us to standardize the quality (e.g. coverage of control genes, average size of transcripts, coverage of target regions).
In addition, we have solved the problem of complexity in library preparation. The three days of work required for preparation of libraries, up to the loading of the pools, have been reduced to only a few steps. The manual procedure now involves the set-up of the automation tool, loading of the pools into the sequencer, and approx. 2–3 days for the analysis of the data (for 16–24 samples).
“[With automation], we have solved the problem of complexity in library preparation. The three days of work required for preparation of libraries have been reduced to only a few steps.”
What are the greatest benefits of using SOPHiA DDM™ ROS?
The greatest benefit of this application is the advanced variant filtering capability and pathogenicity pre-classification that optimizes the data analysis flow and facilitates data interpretation.
In addition, the creation of a particularly exhaustive report for clinical information and ongoing trials relating to the pathologies under examination, make the results usable by the clinician for the therapeutic choice and prognosis of the disease.
We’d like to thank Manuela Badiali, Rita Congiu, and Stefania Murru for their time and for sharing their experience. Click here to learn more about SOPHiA DDM™ RNAtarget Technology and request a demo!
The Laboratory of Genetics and Genomics use a CE-IVD version of SOPHiA DDM™ ROS called SOPHiA DDM™ Dx ROS, available in in the European Economic Area (EEA), the United Kingdom and Switzerland. The CE-IVD application is not designed for use with automation – The Laboratory of Genetics and Genomics have validated the automated workflow for clinical use. SOPHiA GENETICS products are for Research Use Only, not for use in diagnostic procedures unless otherwise specified.
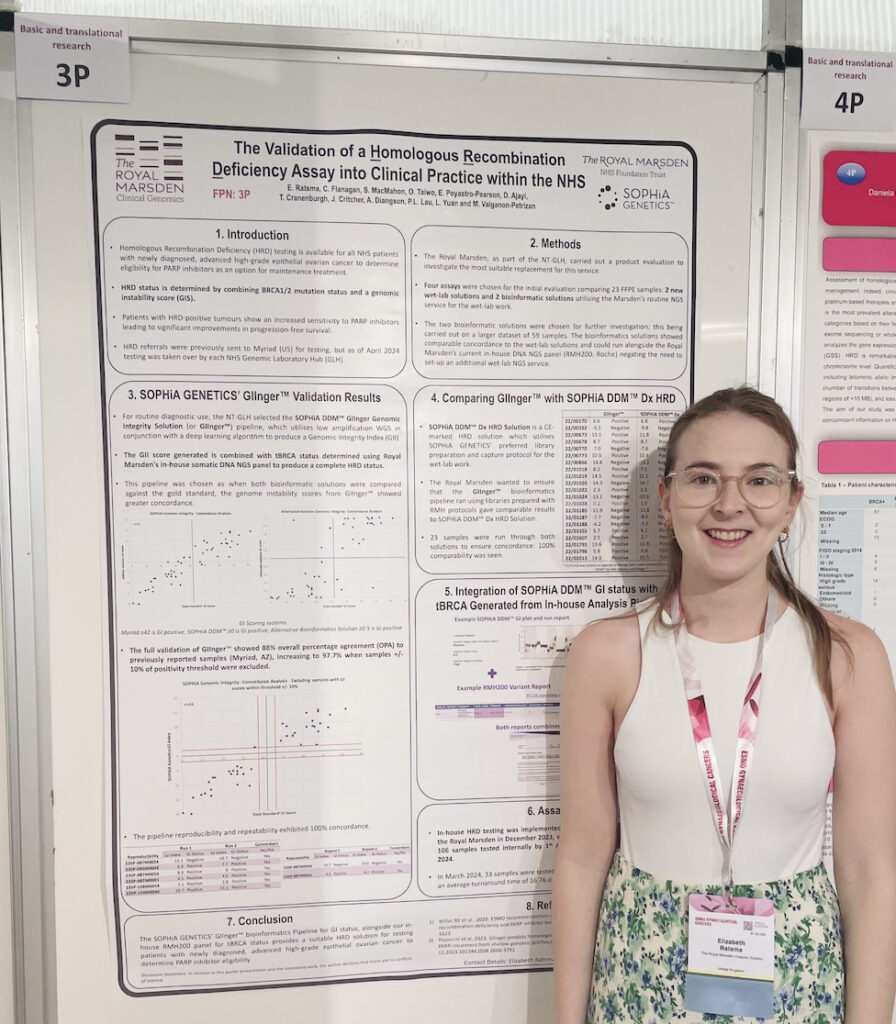
We were delighted to meet with the two presenters of this poster, Elizabeth Ratsma, Pre-Registration Clinical Scientist - Cancer Genomics and Charlotte Flanagan, PhD, Innovation Lead at The Royal Marsden NHS Foundation Trust. The poster was recently presented at the ESMO Gynaecological Cancers Congress 2024 in Florence, Italy.
We would like to warmly thank them for their insightful presentation and for sharing the key takeaways of their study and experience with us through this following interview.
Dear Elizabeth and Charlotte, could you please share with us more information about the scope of this study? What was the aim of it?
Homologous Recombination Deficiency (HRD) testing is available to patients in the NHS who have been newly diagnosed with high-grade epithelial ovarian cancer.
Prior to December 1, 2023, eligible patient samples were sent to the United States for testing via Myriad. The aim of this study was for the North Thames Genomic Laboratory Hub to establish an alternative in-house method for testing ovarian FFPE samples for genomic instability and BRCA mutation status. This would provide an HRD score to enable patients to access PARP inhibitors.
How was your experience with implementing the SOPHiA DDM™ GIInger Genomic Integrity Solution?
We had experience using the SOPHiA DDM™ Platform from previous demos trialing the SOPHiA DDM™ Dx HRD Solution. The SOPHiA DDM™ GIInger Genomic Integrity Solution (or GIInger™) is accessed in a similar process, and it was not difficult to navigate this new workflow in the software. Our bioinformatics team developed a CLC script with support from SOPHiA GENETICS to upload our low-copy WGS sample FASTQ files into the SOPHiA DDM™ Platform, where they are analyzed and genomic instability scores are generated.
SOPHiA GENETICS were receptive to our suggestions to help optimize functionality for use in a clinical diagnostic setting. For example, updating the format and accessibility of reports to facilitate pairing the results with our in-house tBRCA reporting (RMH200, Roche analyzed using DRAGEN, Illumina) and LIMs system.
We have received good technical support when needed through the SOPHiA GENETICS JIRA system.
"We are looking forward to future updates, including a web-based portal and automated download of result files for our clinical scientists to access with ease."
In the scope of this study, you compared GIInger™ and the SOPHiA DDM™ Dx HRD Solution. Could you please summarize the purpose and outcomes of this comparison?
The SOPHiA DDM™ Dx HRD Solution is a CE-marked HRD solution that utilizes SOPHiA GENETICS’ preferred chemistry for whole genome and capture library preparations. At Royal Marsden Hospital (RMH), we had previously implemented a robust NGS protocol (RMH200) capable of producing BRCA capture and low-copy whole genome libraries. To optimize operational efficiency, we decided to explore using our existing automated chemistry and library preparation workflow and opt for a bioinformatics solution to analyze the whole genome data for genomic instability to provide a HRD status.
To this end, 23 samples (previously tested via an orthogonal method) were sequenced using our in-house NGS chemistry and analyzed using GIInger™ paired with our in-house tBRCA calling and compared to the SOPHiA DDM™ Dx HRD Solution, which utilizes different chemistry and a full bioinformatics solution. Reassuringly, we found 100% concordance.
Thank you for sharing these insights with us! To conclude this spotlight, we would like you to share the key takeaways of this study.
Utilizing the SOPHiA GENETICS GIInger™ bioinformatics solution paired with our in-house RMH200 panel for tBRCA status, RMH successfully launched in-house HRD testing in December 2023. By April 1, 2024, we had tested 106 samples internally, achieving recent average turnaround times of less than 21 days.
"The support provided by SOPHiA GENETICS has been sufficient and rapid, which has been invaluable during the first six months of this new service."
We would like to thank Elisabeth and Charlotte for their participation in this spotlight.
Learn more about:
SOPHiA DDM™ GIInger Genomic Integrity Solution
SOPHiA DDM™ Dx Homologous Recombination Deficiency (HRD) Solution
Disclaimer
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise. The SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution is available as CE-IVD product for In Vitro Diagnostic Use in European Economic Area (EEA), the United Kingdom and Switzerland.
Tell us a bit about your role.
I am a Senior Clinical Genomic Variant Analyst at the Broad Clinical Labs – a CLIA/CAP certified lab at Broad Institute of MIT and Harvard. Broad Clinical Labs offers fee-for-service clinical genomic testing, and I am part of the clinical interpretation team for whole genome sequencing. Clinical testing can be ordered from our laboratory by clinicians; our lab performs whole genome sequencing and provides technical genome data and an interpretative report. Our team’s goal is to review the genome data and find reportable variant(s) to share with clinicians.
What type of cases do you work with?
Some of the testing we perform is panel based, but mostly we work with whole genome sequencing data. Because we do not bill insurance, we create relationships with individual institutions or organizations and serve as their sequencing and interpretation partner. Currently, some of the projects we are partnering on include cases focused on rare disease, a CICU cohort, and neurodevelopmental disorders. We use Alamut™ Visual Plus as an add-on tool to our existing variant interpretation workflow when we need additional information about an identified variant.
What are the biggest benefits of Alamut™ Visual Plus that help you overcome your variant interpretation challenges?
Seeing and understanding the genomic context that a variant exists within can be critical for interpretation. Alamut™ Visual Plus makes it easy to see the context at a glance, providing link-outs to other databases for even more detailed information.
Alamut™ Visual Plus is especially helpful when I’m trying to assess the potential impact of splicing. Alamut™ Visual Plus provides calculated splice predictor scores, but I also want to see exactly where in the gene the variant falls in relation to the exon and to visualize the potential impact of exon skipping and whether it’s likely to result in an in-frame or out-of-frame change. Alamut™ Visual Plus makes this easy to visualize.
Can you share an example where this made a difference?
I was recently assessing a splice variant in a gene known to cause a disease that fit with the given phenotype. The variant was +3bp away from a codon, but this alone wasn’t sufficient to make it reportable. After visualizing the splicing impact of the variant within Alamut™ Visual Plus to confirm the potential for out-of-frame exon skipping, I felt more confident to report the variant as a VUS of interest.
Do you have another example where Alamut™ Visual Plus helped you assess a variant?
In another recent case, I had found what looked to be a potentially relevant variant. This variant had been reported using different historical nomenclature in several publications. I came across one paper with a series of cases and variants, that included a variant at both the historic nomenclature and the current nomenclature position. Alamut™ Visual Plus let me easily view and scan the context of the amino acid sequence across different transcripts to appropriately anchor myself to the right reference. This allowed me to confirm I was using correct variant information from this publication in my interpretation.
We’d like to thank Katherine Lafferty for her time and for sharing her experience. We look forward to continuing our conversations with Broad Clinical Labs! Click here to learn more about Alamut™ Visual Plus and request a free trial.
Assessment of Homologous Recombination Deficiency (HRD) and BRCA mutational status in high-grade serous ovarian carcinoma (HGSOC) samples has become crucial for personalized medicine, guiding treatment decisions, and predicting response to specific therapies, such as PARP inhibitors.
The aim of this multicentric study, published at this year’s European Society of Gynaecological Oncology (ESGO) conference was to assess the reliability and reproducibility of SOPHIA DDM™ Dx HRD Solution across three different hospitals (Hospital del Mar, Hospital Vall d’Hebrón and Hospital Clinic de Barcelona) to consider its implementation as a decentralized solution in routine molecular diagnostics.
Our Spanish team led by Giovanni Velotta, CS Manager at SOPHiA GENETICS, was happy to meet Dr. Gardenia Vargas, Molecular Geneticist and Responsible for Hereditary Cancer and Rare Disease Molecular Diagnosis at Hospital del Mar in Barcelona, Spain, who joined us for an insightful interview, sharing her experience in implementing SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution.
Before I answer I would like to thank you and thank AstraZeneca for their support in this study. I must mention that all my answers represent my own perspectives and not necessarily the official stance of the hospitals involved in this project.
The study aimed to investigate the feasibility and effectiveness of implementing the SOPHiA DDM™ Dx HRD (Homologous Recombination Deficiency) Solution within the clinical setting of three hospitals. This includes evaluating various aspects such as the practicality of integrating the test into existing diagnostic workflows, the accuracy and reliability of test results, the impact on patient care and outcomes, and the potential benefits and challenges associated with in-house testing. The study seeks to provide valuable insights into the utility and suitability of adopting the SOPHiA DDM™ Dx HRD (Homologous Recombination Deficiency) Solution as a routine diagnostic tool for identifying HRD in ovarian cancer patients.
First of all, I must say that all three hospitals worked equally on this project, and the idea of doing it was born before I was incorporated to it. Beatriz Bellosillo from Hospital del Mar was the principal investigator, and we had different responsibilities such as protocol writing, ethics approval coordination, securing funding to support the study's activities, and management and allocation of some resources like sequencing supplies.
And of course, it was truly a rewarding experience collaborating with renowned hospitals and esteemed colleagues.
Currently, while we have conducted a multicenter study to evaluate the feasibility of implementing the SOPHiA DDM™ Dx HRD Solution, we have not yet implemented it into routine diagnostic practice at my hospital.
The implementation process requires significant financial support, particularly for acquiring the necessary equipment, reagents, and personnel training. Additionally, to ensure cost-efficiency and timely responses to patients, we need to conduct sequencing runs with a minimum number of samples per run.
Unfortunately, our hospital alone does not have sufficient sample volume to meet this requirement. Therefore, we are actively seeking financial support from government agencies to fund the implementation of the study. Additionally, we are exploring collaborative efforts with other hospitals or healthcare institutions to pool together an adequate number of samples for sequencing runs, thus reducing costs per sample and ensuring a rapid response to patients.
By securing both financial support and an adequate sample volume, we aim to overcome these logistical challenges and proceed with the implementation of the SOPHiA DDM™ Dx HRD Solution into our diagnostic routine, ultimately enhancing our ability to provide efficient and effective patient care.
It highlights the importance of collaboration among hospitals, government agencies, and other stakeholders to support the implementation of advanced diagnostic technologies into routine clinical practice.
Overall, the study provides valuable insights into the challenges and opportunities associated with integrating advanced genomic testing into routine diagnostics, with the aim of improving patient care and outcomes.
This collaborative study concludes that SOPHIA DDM™ Dx HRD Solution provides reliable and consistent results across different hospitals and sequencing runs.
These findings contribute to the growing body of evidence supporting the use of the decentralized SOPHiA DDM™ Dx HRD Solution in clinical settings for genomic analysis.
DISCLAIMER: SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise. SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution is available as CE-IVD product for In Vitro Diagnostic Use in Europe and Turkey.
View the video
The term « SOPHIA » used by the speaker refers to SOPHiA GENETICS and its products. The opinions expressed during this presentation are these of the speaker and may not represent the opinions of SOPHiA GENETICS. SOPHiA GENETICS does not provide support in the validation of custom products for clinical use. SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution is available as a CE-IVD product for In Vitro Diagnostic Use in Europe and Turkey. SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
Marinela Lekka
Field Marketing Lead - EMEA, SOPHiA GENETICS
mleka@sophiagenetics.com
Dr Alessandra Terracciano is part of the large team at the Medical Genetics Laboratory in the Bambino Gesù Children’s Hospital, Rome, Italy. She analyzes next-generation sequencing (NGS) data, with a focus on rare pediatric conditions. Her laboratory primarily works with samples from the Bambino Gesù Children’s Hospital across multiple specialties, but it also acts as an Italian reference center for some conditions, such as kidney diseases.
Dr Terracciano kicked off our symposium at the European Society of Human Genetics (ESHG) Conference 2022 by describing the laboratory workflows, tailored sequencing analytics, and multidisciplinary team meetings that enable the Bambino Gesù Children's Hospital to successfully solve challenging pediatric cases. Afterwards, she kindly sat down with us to answer a few questions on her experience working with SOPHiA GENETICS and what she sees for our collaboration in the future.
Tell us a little bit about your collaborations with SOPHiA GENETICS
My experience with SOPHiA GENETICS started when developing the SOPHiA DDM™ Platform Nephropathies Solution. We collaborated to develop an enrichment kit and analytical pipeline that successfully discriminates the PKD1 gene from its pseudogene. Although it was a tricky request, by the end of the validation process, the performance of the application was very, very good! For more details about this collaboration, read the case study with Dr Antonio Novelli
I also worked with SOPHiA GENETICS on a validation program for the analysis of genes associated with metabolic diseases. Again, SOPHiA DDM™ Platform was very helpful, as the solution is very robust and able to detect point mutations and copy number variations (CNVs) at the same time.
SOPHiA DDM™ Platform is very user-friendly - the visualization makes it easy to analyze CNVs.
For whole-exome sequencing (WES), our lab performs a double-analysis. We use both our sequencer’s pipeline, and SOPHiA DDM™ Platform, which is particularly useful for tricky cases such as those involving CNVs.
What are your favorite SOPHiA DDM™ Platform features?
SOPHiA DDM™ Platform is very robust. I trust it, believe the data, and feel safe when using it. In addition, the platform is very user-friendly, with the visualization making it easy to analyze CNVs. It is also helpful that I can easily export data from the platform, for further analysis using other tools if necessary. The platform is very comprehensive. If I find, for example, a point mutation, I have access to all the relevant information about it, such as the ClinVar annotation, the frequency, and the experience of other users. I can tag variants myself and can see the tags made by any of my colleagues.
The platform is very comprehensive – I have access to all of the relevant information about a point mutation, for example
In your opinion, what role does SOPHiA GENETICS play in improving rare and inherited disease research in the future?
SOPHiA GENETICS have a very powerful tool for people to share their experience. Sharing information on NGS analysis for rare pediatric conditions is very important, especially for the interpretation of variants of uncertain significance (VUS).
SOPHiA DDM™ Platform is a very powerful tool for people to share their experience
We’d like to thank Dr Terracciano for her time and all her work. We look forward to continuing our successful collaboration with the Bambino Gesù Children’s Hospital.
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us at support@sophiagenetics.com to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.