Menu
Menu
Just two decades ago, cancer was largely considered an organ-based disease. For example, lung cancer, despite having known histological subtypes, was uniformly treated as a single disease – chemotherapy for all. Today, advances in clinical genomics have transformed lung cancer into a collection of rare diseases defined by a long tail of distinct genomic alterations. Building on this knowledge, targeted therapies have gradually improved patient outcomes for eligible patients. However, significant medical needs remain, particularly regarding overall survival (OS) or achieving durable cures. Most lung cancer patients in first-line therapy do not have a clear molecularly driven cause and almost uniformly receive an immunotherapy-based treatment as a standard of care. Despite massive investments, single-biomarker approaches have failed to reliably predict response to immunotherapy, leaving clinicians unable to determine which patients will benefit most.
To address these challenges, precision medicine must shift from a siloed, single-biomarker approach to a more integrated multimodal approach, combining genomic, imaging, clinical, and biological data. Intuitively, taking a more holistic view of the patient, the tumor, and the host environment should open a stronger window into the biology of health and disease. Yet, realizing the full potential of this approach requires a transformation of the underlying data infrastructure. This includes breaking down silos across data modalities, standardizing and harmonizing datasets and promoting real-world knowledge sharing across institutions. Such transformation is necessary to unlock the power of artificial intelligence (AI) applied to multimodal healthcare data.
In this article, we explore the groundbreaking potential of multimodal AI-driven technology as a key driver of a new era in precision medicine. This paradigm shift promises not only to accelerate innovation but also to improve patient outcomes and expand equitable access to care. With competition intensifying and regulatory scrutiny increasing, biopharma companies must embrace this transformation to stay ahead and reshape the future of precision oncology.
Over the past decade, we have seen a dual revolution in healthcare: the explosion of multiple types of digital health data being produced at scale in clinical routine (e.g., genomics, imaging, electronic health records (EHR) entries) and generational breakthroughs in analytics capabilities (e.g., machine learning, deep learning, foundational models). In theory, this combination should have unlocked the full potential of precision medicine, but in practice, we are arguably still at its Stone Age. One of the root causes of this lies in the fact that healthcare data remains largely fragmented and unharmonized, and the tools to integrate and harness it effectively are often lacking. Additionally, there is no built-in incentive in the ecosystem for large-scale data and insights sharing across institutions, while preserving privacy. The “publish or perish” mantra in academia still tends to encourage data hoarding — consciously or not — further hindering collaboration.
Similarly, on the biopharma side, we see increasing interest and expertise for AI-driven approaches on specific data modalities (e.g., digital pathology, radiology). Yet, these initiatives are still rarely connected in a truly multimodal framework. Proprietary clinical trial databases remain challenging to harmonize and merge together due to heterogeneous patient consents, compliance issues, and required investments.
A fair question to ask is, therefore—is it even worth the trouble?
Although we are just beginning to tap into the capabilities of multimodal AI-driven technology, it is already driving significant advances in our understanding of health and disease. By seamlessly integrating and analyzing diverse data sources, this approach enables a more holistic perspective of complex diseases like cancer, and the patient beyond the disease.
The practical application of AI-powered multimodal technology can help biopharma companies solve complicated biological puzzles and overall optimize the drug development process (Figure 1).
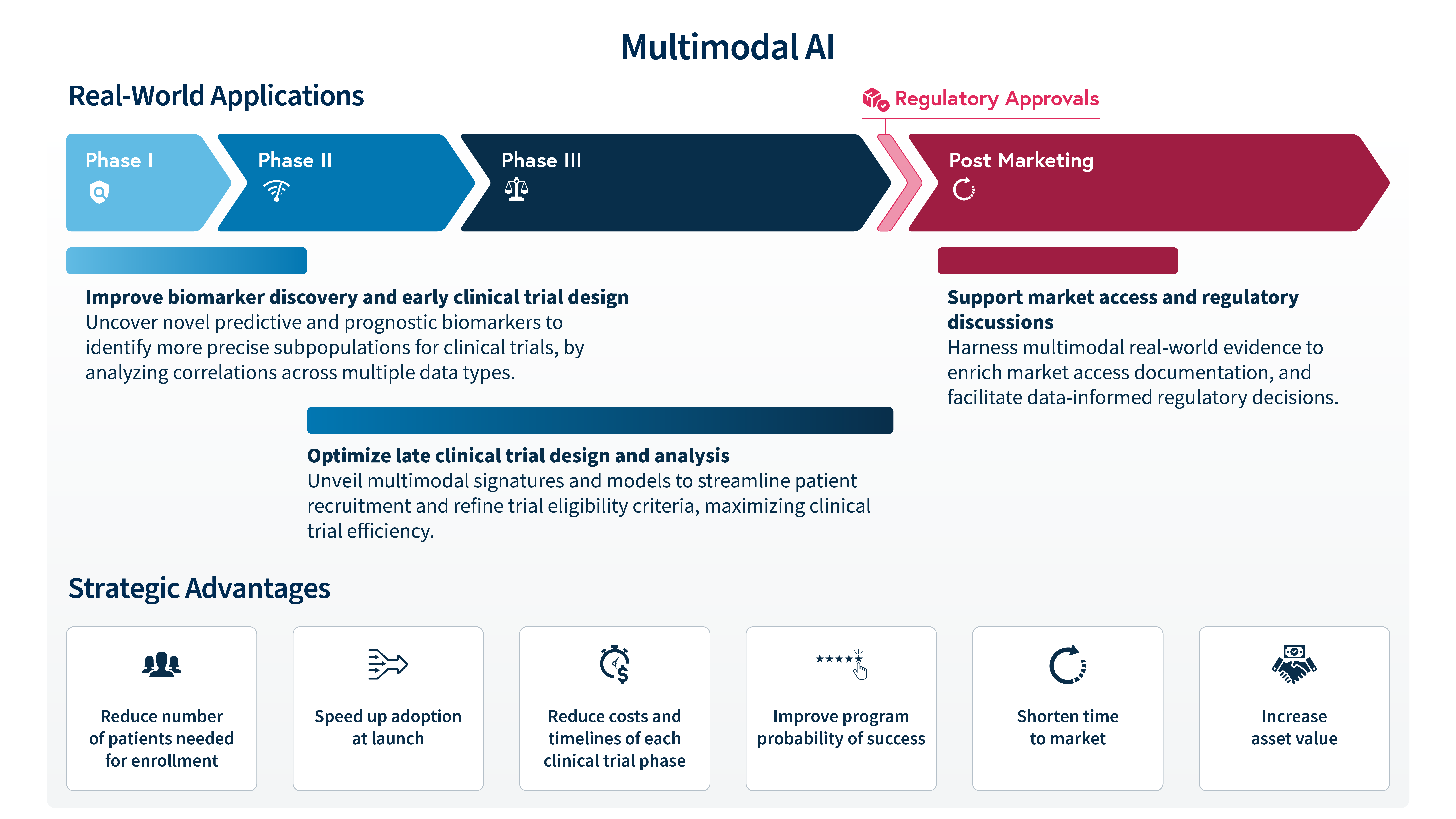
At SOPHiA GENETICS we believe that multimodal AI is no longer an option but a necessity to accelerate precision medicine. Our cloud-based SOPHiA DDM™ Platform seamlessly integrates and standardizes diverse data types into a unified analytical framework, comprising state-of-the-art specialized computational modules for data processing and analysis (e.g., genomics, radiomics), including a dedicated multimodal factory. This engine combines, extracts, and structures complex multimodal data to fuel the development of predictive analytics, delivering actionable insights that empower data-driven decision-making through an intuitive, user-friendly interface (Figure 2).

The potential of this multimodal approach is evident in the initiatives we are leading here at SOPHiA GENETICS. One noteworthy example is the TRIDENT project, presented at ESMO 2024 (Skoulidis et al., 2024). TRIDENT was a retrospective multimodal re-analysis of AstraZeneca’s Phase 3 POSEIDON trial (NCT03164616). AI-powered predictive models of treatment benefit were trained on the totality of the clinical trial data, including clinical, biological, imaging and genomics data, with the intent to identify patient subpopulations that may derive greater benefit from the addition of a CTLA-4 inhibitor on top of a PD-L1 and chemotherapy backbone in first-line non-small cell lung cancer. These models yielded signatures identifying approximately 50% of the trial population in scope that would be predicted to benefit from the addition of CTLA-4 inhibition, with a hazard ratio reduction from 0.88 (95% CI, 0.68-1.12) to 0.56 (95% CI, 0.33-0.97) in the non-squamous histology population (Figure 3). These multimodal signatures are clinically interpretable and can be readily deployed in the real-world setting on the SOPHiA DDM™ Platform for further clinical research.
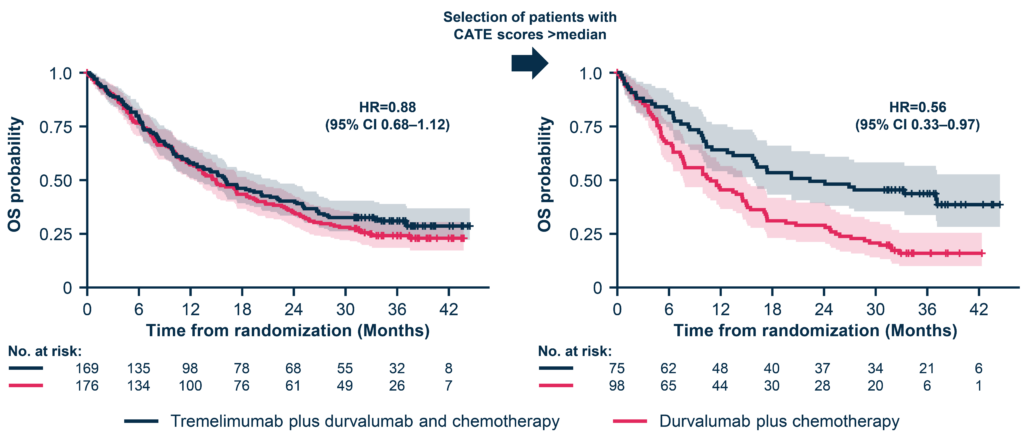
This proof-of-concept analysis highlights a fundamental observation: traditional methods of analyzing and making sense of existing data, for example, through univariate or multivariate analyses, can leave significant portions of the full picture unseen. In contrast, new multimodal approaches have the potential to reveal insights that would otherwise remain hidden.
The transformational findings from this lung cancer project are not an exception. Similar results were obtained in other cancer types, such as in kidney cancer (Boulenger de Hauteclocque et al., 2023; Margue et al., 2024), and breast cancer (Groheux et al., 2025).
To successfully implement a multimodal AI-driven strategy, it is essential to begin with a clearly defined clinical question. What are you trying to predict or stratify, and why does it matter clinically? What is your endpoint of interest? These considerations will dictate the type, quality and volume of data required for a successful analysis.
A truly multimodal model demands a deep understanding of the signal-to-noise ratio within each individual data modality. For example, in genomics: do you know how the DNA of the tumor was sequenced, which chemistry was used, and on which sequencing platform? How was the variant calling performed, and what are the known limitations? Only with this level of detail can you distinguish meaningful biological signals from background noise. The same applies to radiology, where harmonizing image data across different platforms and reconstruction techniques is essential. Skipping these foundational data preparation steps risks identifying patterns in noise rather than signal.
Once the dataset is well understood and curated, data augmentation can be applied—for instance, through radiomics analysis of 3D medical imaging or pathway analysis of relevant genomic variants.
After augmentation, data aggregation becomes the next critical step. This involves integrating diverse data types into a unified analytical framework. From here, various statistical learning methods can be selected based on the clinical objective. Typically, imputation techniques for missing data need to take place before feature selection, statistical model selection, and calibration. Finally, ensuring analysis interpretability, both at the cohort and individual level is an important step in facilitating discussions with oncologists and other healthcare professionals concerning the methodology and the outcomes of the models (Figure 4). This can be achieved by using traditional machine learning models.
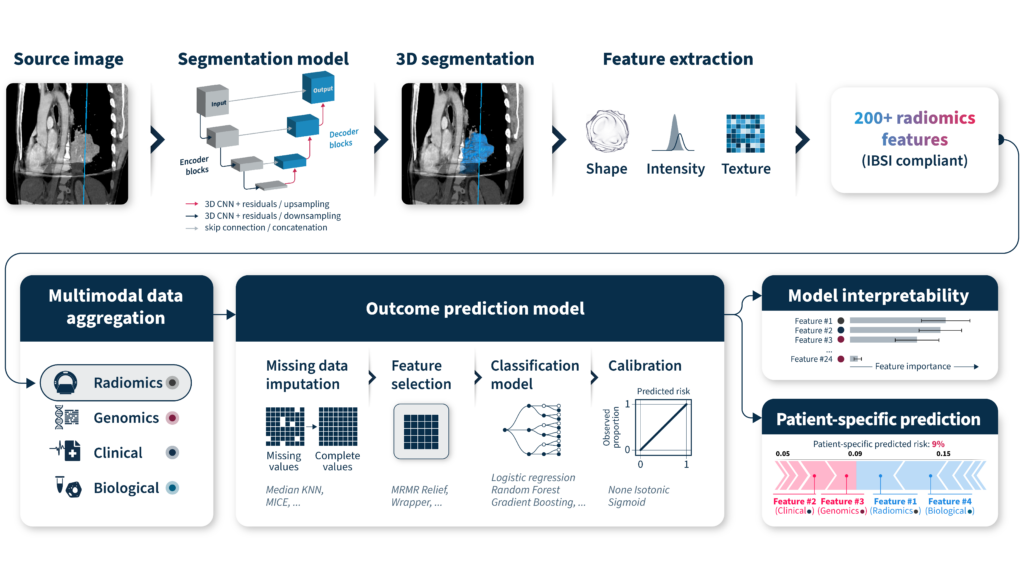
An often overlooked yet crucial step is the deployment of the models in a real-world setting. How will end users interact with the model? How will data be input and managed? What safeguards are in place for data privacy and security, and how will computational infrastructure scale?
At SOPHiA GENETICS, we believe that the cloud-native SOPHiA DDM™ Platform is uniquely positioned to spearhead this movement. Already adopted by over 800 healthcare institutions across more than 70 countries, the platform has securely processed data from more than 2 million patients, ensuring privacy while enabling impactful, AI-powered multimodal insights at scale.
The transition from single-modality to multimodal AI-driven analysis represents a paradigm shift in precision medicine. Organizations that successfully integrate diverse data modalities and multimodal technology will be best positioned to drive better patient outcomes and maximize drug development success.
Realizing this potential, however, demands more than technical innovation. It demands systemic transformation across the overall healthcare landscape — from evolving regulatory frameworks for multimodal CDx, to updated reimbursement models, standardized deployment practices— inclusive of post-market surveillance—, and greater education for both clinicians and patients.
For biopharma companies, the path forward is clear:
About twenty years ago, cancer was still considered an organ disease. Looking back today, this may look like distant, medieval times. Twenty years from now, new generations of life sciences professionals may look at 2025 in a disturbingly similar way. The multimodal revolution is only now getting started.
Written by Philippe Menu, MD, PhD, MBA - EVP, Chief Medical Officer & Chief Product Officer, SOPHiA GENETICS
References
Skoulidis F et al. 1325P TRIDENT: Machine learning (ML) multimodal signatures to identify patients that would benefit most from Tremelimumab (T) addition to durvalumab (D) + chemotherapy (CT) with data from the POSEIDON trial. Ann. Oncol. 35, S842–S843 (2024).
Boulenger de Hauteclocque A et al. Machine-learning approach for prediction of pT3a upstaging and outcomes of localizaed renal cell carcinoma (UroCCR-15). BJU Int. 2023; 132(2):160-169. doi: 10.1111/bju.15959.
Margue G et al. UroPredict: Machine learning model on real-world data for prediction of kidney cancer recurrence (UroCCR-120). NPJ Precis Oncol. 2024; 8(1):45. doi: 10.1038/s41698-024-00532-x.
Groheux D et al. FDG-PET/CT and Multimodal Machine Learning Model Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Cancers. 2025; 17(7):1249. doi: 10.3390/cancers17071249.
We sat down with Prof. Jacques Cadranel, International Coordinator of the DEEP-Lung-IV study and Head of the Pneumology Department at the Hospital Group University Hospitals of Eastern Paris, who shared his experience in this collaboration with SOPHiA GENETICS, and the importance of the integration of multimodal data in clinical practice to advance personalized medicine in lung cancer.
Watch the spotlight:
Hello Professor Cadranel, thank you for welcoming us today at the Tenon Hospital. Could you explain to us what DEEP-Lung-IV is and the genesis of such a project?
The genesis of this project is 2020, so it's been a while. With the desire to move from a slightly comparative medicine of treatment arm to arm, to a more individualized medicine.
Taking into account the fact that we have what we call artificial intelligence (AI), which allows us to accumulate a lot of data, and ultimately be able to develop signatures that we cannot develop in our heads, or even with, a usual statistical approach. And also to have the impression that the patient cannot be reduced to a sex, a weight, a performance status, an imaging test, or a molecular test.
The patient is a whole, and as long as we haven't integrated this whole, I think we will still be a long way from individualized medicine.
Could you explain the DEEP-Lung-IV project as it is today?
At the time, and what it has become, was first to say: “Let’s do something from the actual standard of care as it exists today”. That is to say, not to build a project that won't be applicable in real life. That was point number one.
Point number two was to accumulate the usual clinical data we have in our medical records, the usual biological data, blood counts, creatinine levels, liver tests, and imaging data while avoiding focusing solely on what we call the targets that we measure in interventional trials, but rather taking into account the patient as a whole. So, having a radiological phenotype. And then also integrating molecular parameters. Creating an initial profile of the patient, treating them as they were treated, and having their (outcome) information at the first assessment – are they stable or responders? - so that we can subsequently create signatures that would allow us to define their treatment outcomes before exposure.
From that, we can predict not only what treatment the patient received but what they would have been like if they had received another treatment. So that's the first step, that's the basis of DEEP-Lung-IV.
We talk about predictors, we talk about making the signature available so that it can be deployed in the clinical routine. What does such a platform look like to you?
The platform would be similar to what SOPHiA GENETICS was kind enough to show us. It's a platform that is, first and foremost, very tactile, very easy, very pleasant.
That's essential in the Multidisciplinary team meetings (MTBs). We can't get carried away, it musn’t be complex and should be extremely user-friendly. What we envision is a simple interface that provides some kind of a detailed patient profile, including characteristics such as pathology, performance status, and offering clear probabilities for treatment response, progression, overall survival, and for each therapeutic option.
Why choose SOPHiA GENETICS as a partner for such a project?
I believe that’s not how you find a partner. A partner, you look him in the eye and you say, "I want to see". And then you start exchanging, and you want to see even more. And little by little, that's how you build a bond. I wouldn't have been able to choose SOPHiA GENETICS if I hadn't felt, from the start, the idea that SOPHiA GENETICS was coming towards us with was really important, because that's not what usually happens. Meaning that we have private partners who do not listen to the creativity of the doctors and of their understanding of the complexity of the patients, so that they can provide us with tools that respond to this complexity. Usually, these partners come with their prototypes and expect us to agree with it.
This is what I really appreciated about SOPHiA GENETICS. It was a real partnership from the beginning. Together, we created something very original, something that neither of us could have done alone. And that's what's so enjoyable about this collaboration.
Professor Cadranel, you are the international coordinator of the study and also the scientific committee's chair. Could you tell us what this represents for you, in terms of challenges and opportunities, and why you accepted such an appointment?
Why did I accept? Because I share this baby with SOPHiA GENETICS. It's also my baby, and it's a really meaningful project for me. I believe that conceptually, we can completely change the paradigm and move on from a Newtonian medicine, that is: we observe A, which does better than B in interventional trials. And then we happily apply this, trusting the hazard ratio to double the probability of response, etc. But ultimately, that's not how it works. It's useless, and we should rather lean towards what I call a kind of quantum medicine. That's why I find this project extremely exhilarating and exciting, and I was given some chance to be part of it. That's also unique. That's why I don't want to miss out.
So the challenge is getting others on board. Right now, SOPHiA GENETICS and I, we need to accelerate the project a little more to produce results. And we are close. We hope that this new year will be the one of results and of raising awareness of this project. And SOPHiA GENETICS may not yet be aware that doctors aren't ready to hear what we're going to tell them, and so there will be some challenges ahead of us. We need to educate the doctors currently conducting the investigation because I think they didn't fully understand what they were getting into. They provided data for this study, they trusted SOPHiA GENETICS and also me, but I think they didn't understand what we were going to offer them at the end. And this is an exceptional challenge because it truly represents a complete paradigm shift for precision medicine.
Professor Cadranel, could you explain to me what you expect from the multimodal approach and the use of AI in clinical routine?
When we discussed DEEP-Lung-IV early on, I represented a little slide with my brain, which was composed of the performance status, the burden of the disease, molecular biology, organ pathologies, extent and type of metastases, and so on. And I told you, in fact, I integrate all of this unconsciously, but I also integrate it consciously.
There's still a need for us, first of all, to ultimately try to mimic our approach, without us realizing it, is what I call the black box, to make things more objective and also a little more reproducible, in particular with some “grey” situations where we don't really know what the best strategy is for the patient. If we resubmit the same patient, changing their name, changing their sex, or gaining one kilo, we realize that halve the times, we don't give the same answer. That's still a problem.
It's a concern that we're not reproducible within a MTB, but between MTBs. So that also means that there's an inequality when it comes to patient care, which we hope will be mitigated once we have that AI signature.
What other applications would you see in the context of multimodality and artificial intelligence?
There are. One part that we don't control is the patient's side. Having a perspective from the patient’s and caregiver’s perspectives. Integrating their perspectives into these signatures would be extremely valuable. Even with the greatest desire to do well as a doctor, when you make an announcement to a patient, when you offer them a treatment, you project an extraordinary amount of information on what the patient will receive and how they will experience it.
We did some sociological work to see how they felt about it. The most enthusiastic, compassionate doctor, the one who wants to do good the most, is wrong in 99% of cases by the patient. Patients are an essential element to consider.
What we should also integrate into this therapeutic signature is how it integrates into people's emotional, social, and personal lives.
We thank Prof. Cadranel for his time and for sharing his experience. To learn more about the DEEP-Lung-IV Study, visit the dedicated page .
SOPHiA GENETICS products are for Research Use Only, not for use in diagnostic procedures unless otherwise specified.
We met with Dr. Sébastien Couraud, DEEP-Lung-IV Scientific Committee member and Head of the Pulmonology and Thoracic Oncology Department at Hospices Civils de Lyon, to talk about his participation in SOPHiA GENETICS’s DEEP-Lung-IV study and reflect on the benefits of multimodal approaches to transform precision medicine and improve patient outcomes.
Watch the spotlight:
Hello Sébastien, thank you for receiving us here at Lyon Civil Hospitals (HCL). You are the biggest recruiter of the DEEP-Lung-IV study that we launched a few years ago and also a member of the scientific committee. I would like to have your perspective on why you joined the study, and then your vision on the project in general.
Hello Marion, thank you very much for the invitation. I am very happy to welcome you here and to have a discussion on the DEEP-Lung-IV project.
We were immediately won over by the ambition of the project and by the fact that this project was very multimodal, precisely.
And I think that is really what we are going to discuss together today. This very multimodal, very ambitious side immediately won us over. In addition, it is true that we have had quite a few strong relationships with the members of your team from the start. We already knew each other before, so it was quite logical for us to finally support you on this new project.
Could you tell us more about the objective of the DEEP-Lung-IV study and how this study will meet the objectives of precision medicine in the future?
The objective of the DEEP-Lung-IV study is really to go, collect quite a massive amount of data on patients who are treated in different investigation centers and who are treated for lung cancer.
And in fact, the principle is really the multimodal collection of massive data, to then be able to create decision support tools that will help us on a daily basis.
Concretely, it manifests itself in a quite simple way - in reality, when we take care of a new patient, these are patients where we will ultimately integrate all of the data that we have generated for this patient. And collected in a database.
The multimodality here comes from the fact that we will collect radiology, pathology, and molecular biology data, and connect it with clinical data.
All this data will ultimately make a very large database, with centers from all over the world, and will then allow us to ultimately generate decision-making tools.
For the future, how do you see the next steps of the study and the collaboration with SOPHiA GENETICS?
For the future, obviously, the first step is the results. We were talking about it earlier, we need to have these results, see precisely the type of results that we have, the tools that have been generated, and we will then have to ask ourselves the question of whether we can use and integrate these generated tools into practice, how to do it and evaluate it.
We are finally at the beginning of a collaboration, and it would be a shame to stop on such a good trajectory. The objective is to continue the collaboration with SOPHiA GENETICS because it is indeed really important, from now on, to enter a partnership a little more operational, if I dare say.
You previously told us that artificial intelligence (AI) was expected in real routine practice. In your opinion, what would allow us to bring this to routine?
This step towards the clinical routine of integrating AI, that really is a very good question and I think it's really, if I may say, the golden question to which ultimately no one really has an answer today. We all think that AI will have a strong impact on medicine, in several dimensions of medicine. Obviously, when we talk about lung cancer, the decision of which today is very multimodal, and is a field of knowledge that is expanding almost hour by hour.
In reality, we imagine that AI will be a very important decision-making tool, daily. Nevertheless, we work on humans and we work with patients, with lives. So everything we do must necessarily be evaluated and we must be certain of what we do.
And that's the important element and what we're missing today. And what we're missing today are studies that will allow us to show that compared to the absence of artificial intelligence, the addition of it improves patient care on very specific events, such as survival, progression-free survival, treatment tolerance, the choice of a more suitable treatment, etc. So really the next step is prospective evaluation studies that will allow us to integrate these tools into real life and compare them to the outcome and current care.
Sébastien, let's think about the first day of the launch of the project and, with hindsight over these 4 years. If we had to do it again, would you join in?
That's a good question. Yeah, I think so. Yes, I think there was a bit of a crazy side indeed when you came to see me and you told me “We are going to take all the data from your patients, integrate them, and you're going to send everything to us”. It still required a lot of organization for us, but finally, I think we would do it again the same way. The collaboration was really pleasant in reality, that is to say, that it was done in a fairly simple, fairly flexible way. And in reality, with a little bit of organization on our side. Objectively, it went well, so I think that I would do it again. Yeah, I would sign again.
We have been working together, HCL and SOPHiA GENETICS, for several years. Could you tell us about the perception you have of SOPHiA GENETICS as a company?
Yes, so it's true that we've been working together for years.
I think that SOPHiA GENETICS is one of those companies that has entered the health ecosystem through one end and has succeeded in developing this multimodal aspect precisely.
That is to say that when we met at the very beginning, you were really in biology, in genetics. And I remember conversations that date back a very long time. You managed to really open up your field of possibilities by integrating this notion of multimodality and by very quickly understanding the interest of multiple modalities, instead of staying in a single field, in which you were nevertheless quite an expert, but to take a risk by exploring other fields and opening up to other possibilities. And I find that this risk-taking is interesting.
I find it interesting because it ultimately makes it a company that was able to understand a little in advance the interest of multimodal, to bet on it, and today, to open up to it very widely.
So, there is really a fairly innovative side to your company.
Sébastien, today, the treatment decisions for a patient are processed in Multidisciplinary team meetings (MTBs). What do you expect from an algorithm and the machine learning tool in general?
In fact, at the risk of surprising you, the idea is to perhaps be a little less human. You have to understand that today when we take care of a patient, we take care of them based on our instinct, we take care of them based on their story - the patient's story - we know them, we know their story. And then we have our experience - the experience of whether we have other similar patients or not, that we have taken care of. And so that's what ultimately constitutes the decision that we're going to make in the MTB.
It's obviously science, right? There's no doubt about it. We have guidelines, and we rely on these guidelines, but then it's ultimately a collective of clinical experiences, good and bad, that will allow us all together to make the decision that we think is most appropriate for the patient.
Certainly, it's good since it's been working like that for years, and today, we're still practicing good medicine.
However, having a tool that is completely dehumanized will allow us to humanize this question less.
Now, what I'm saying is going to seem very odd, but in fact, it will allow us to tell ourselves that science in this situation, specifically for this group of patients, is telling you that. The human will then come and modulate the scientific decision and will say: the machine's decision is this. I will adapt it with the knowledge of my patient, but at least I will start from something very current, very factual, very scientific, and I will articulate based on it. This is perhaps where it will change things a little.
In practice, this machine support, how do you view it in MTBs? What does it look like?
It looks like a very intuitive interface. In fact, we must not forget that we still have a lot of work. We are increasingly solicited, we are asked for more and more things. The MTBs are becoming very, very heavy. I think all colleagues see that the MTBs are becoming more and more complicated, there are more and more patients. Patients are surviving more and more, which is an excellent thing, but it obviously increases the volume of MTBs.
All this requires us to be very simple and very pragmatic.
The idea is really to have a tool at the service of the clinician, a very intuitive, very easy-to-handle tool, that will give very visible results, very simple for the whole room in a few seconds - I enter the clinical characteristics of my patient, I immediately visualize the data in the database and it will very easily help me to be able to make decisions.
What is also important is that we can vary the parameters a little because sometimes we will hesitate between two strategies. We will say to ourselves: “Here, do I use this molecule or that molecule? Or do I use this diagram or that diagram?” and that finally can be done in one click. Add or remove a therapeutic option and immediately visualize the effect it can have in a cohort. That will help us greatly in our decision making.
We thank Dr. Couraud for his time and for sharing his experience. To learn more about the DEEP-Lung-IV Study, visit the dedicated page .
SOPHiA GENETICS products are for Research Use Only, not for use in diagnostic procedures unless otherwise specified.
We sat down with Prof. Jean-Christophe Bernhard, UroCCR Coordinator, and Dr. Gaëlle Margue, Urology Fellow, at University Hospital Bordeaux, to discuss the collaboration between UroCCR – the French Kidney Cancer Research Network - and SOPHiA GENETICS, and get their insights on the use of AI-powered multimodal approaches to improve patient care.
Watch the spotlight:
Gaëlle, Jean-Christophe, hello. It's a real pleasure to be with you today to discuss the collaboration between SOPHiA GENETICS and UroCCR.
Before we go into more details about this collaboration, I would like you to tell us a little about your background, your life as a surgeon at the hospital, and also introduce the UroCCR network and the role you play within this network.
Gaëlle Margue, I am a junior doctor in urology at the Bordeaux University Hospital. I arrived in 2018 to start my internship and now, I have been a PhD student for two years and my science thesis focuses on kidney cancer, surgical and oncological themes, within the framework of the UroCCR network, the French Kidney Cancer Research Network, and the I.CaRe team, a Kidney Cancer Research team in Bordeaux.
Jean-Christophe Bernard, I am a professor of urology at the Bordeaux University Hospital, and I coordinate the I.CaRe team, the Kidney Cancer Research and Innovation Program in Bordeaux. I am also the national coordinator of the French Kidney Cancer Research Network, UroCCR.
Gaëlle, we worked together on your medical thesis around UroPredict. Could you describe UroPredict, and what it does?
Yes, absolutely. That was the subject of my medical thesis. The initial idea was to better characterize the risk of recurrence after kidney cancer surgery. Therefore, for patients with localized kidney tumors that are operated and are considered cured or in remission after surgery, we want to better determine what factors lead to patient relapse or not. We have prognostic scores to try to determine that, but they are not very effective, and so we wanted to better characterize that, to propose a follow-up schedule tailored to the risk of recurrence, or additional treatments for patients who have a high risk of recurrence.
In UroCCR, we have a lot of data that relates to these kidney cancer surgeries - clinical, biological, imaging, surgical, and monitoring data -, which we were not able to leverage, with traditional statistics, to better characterize the risk of recurrence. So the goal was to create a new machine learning score, a tool to predict recurrence in these patients, based on all this data that we have in UroCCR.
It's quite fascinating, from an outside perspective, to see surgeons saying that we need Artificial Intelligence and Machine Learning, to be able to advance our care. Jean-Christophe, from a strategic point of view, how do you see the collaboration with SOPHiA GENETICS, in particular? And then more broadly, the role these precision medicine tools are expected to play in the future?
We see it as something that will be decisive, and I would say, necessary to maximize the value of all the data we gathered over time.
UroCCR, to go back a little into the history, is a project that dates back to 2006, which was certified by the National Cancer Institute in 2011. It was initially deployed as a multicenter project in 2013 across 11 centers. And today, we’ve grown from 11 centers to 54 and soon 58. And so, the positive excitement of the system means that we are constantly collecting highly qualified data on the pathology of patients who are diagnosed with a urological tumor. We collect this data regardless of the treatment method, whether the patient is in active surveillance, whether he is operated on, whether he is treated systemically with medication, or whether he is treated with interventional radiology.
In doing so, coupled with the increase in the number of centers, and the increase in the number of patients, we reached the 20,000 patients included in the UroCCR, this dataset became even more considerable since we are linked to the SNDS, the National Health Data System, and therefore we can also do medico-economic evaluation, representing nearly 10 million data points available on patients treated contemporaneously by French teams for kidney cancer.
It actually becomes a need, not so much a vision but ultimately a necessity to have new statistical methods to process this data, and derive insights to enhance our ability to provide patients with care that is as personalized and individualized as possible.
This is, I believe, the foundation of our collaboration with you and your team at SOPHiA GENETICS, to explore how we can take advantage of all the data patients have entrusted us with, regarding their illness, since everything is done with the patients’ consent - and I think it’s also important to point this out. Being able to produce new tools, and what Gaëlle said is that her thesis work, which initially is a scientific and fairly general work, I would say, has nevertheless led to the online publication of a calculator, which can now be used in a pragmatic way to answer a question that we may ask ourselves for a given patient.
Ok. So, beyond the research aspect that we were able to carry out together, which I think we are all very happy with, there is this role that an industrial company can play afterward, which will be the deployment of the tools, their validation, their improvement, the entire life cycle of the software.
Are these the things that are beginning to resonate in the minds of the medical profession, or are we still at the beginning and still at a time when everyone needs to find their role?
Yes, it is... It is very timely, because the UroCCR network has just been certified by the ANR, the National Research Agency, as a clinical investigation network on medical devices. These are also themes that we are addressing within the framework of the I.CaRe program, within the framework of the RHU (University Hospital Research in Health) where there is this desire for collaboration between academics and manufacturers for carrying out projects and arriving at outlets that are tools, products that can be used in everyday life and in the routine of patient care.
The UroPredict tool that we co-developed during your thesis, Gaëlle, it is now deployed and accessible. How is it being used by you?
It is a tool that is not yet a certified medical device, so it cannot yet be used to change patient care strategies.
However, it is very useful for discussing with the patient. We can present it as a research tool that helps us better assess their risk of recurrence and concretely show the patient their risk factors for recurrence and their personalized risk. And this allows us to develop the discussion around monitoring or potential complementary treatments to surgery, for the continuation of their care.
Do you see among your colleagues or among the pharmaceutical companies, a little reluctance to apply them, to trust them, at the level of clinical routine?
So, reluctance… I don’t think it’s really a reluctance. I think that at each time there is something new, that potentially can bring a change in practices. There is always an observation phase and we have to assimilate what this novelty can be and what it can bring. We experienced the same thing, for example, for the introduction of robotic assistance in surgery, which today everyone is convinced on its benefit, both for health professionals, for surgeons, but also for patients. There was a whole phase where the community asked itself the question of whether introducing robotics would actually make it possible to do better than conventional techniques that were, I would say, well known, and had been validated for many years.
So there is always this moment where we ask the question of whether it is a real innovation, whether it will really bring added value. There is always this observation phase. Afterwards, I think that more and more, the medical and surgical community is convinced that with technological progress, we are finally able to improve what our daily routine is, what our practice is and obtain additional information that will enrich our practice and patient care.
What do you think the next algorithm we should develop is?
We have several ideas. We could further enrich this one by perhaps incorporating radiomics or pathomics, as we discussed, adding imaging data to increase the precision of the current tool. Then, we could explore many other algorithms with different objectives, such as predicting kidney function loss after kidney cancer surgery, or predicting the risk of morbidity, and intra or post-operative complications.
Objectives and things that we are trying to better characterize and there are several that we could look into and probably we have all the necessary data from UroCCR, we need to be able to exploit it.
And I think if I can complete.
There is also a question that we should address. Every time, we evaluate outcomes that are very objective, focused rather on the practitioner, the surgeon, the quality of his surgery, the outcome, the evolution of the disease, etc.
But I think that we should also be able to work on outcomes that are reported to us by the patient.
Quality of life, satisfaction, anxiety, which are things that we capture in UroCCR and in particular thanks to the UroConnect application. Today, in UroCCR, we have what we call PROMS and PREMS, which are patient-reported outcomes, and I believe this is another field worth investigating, moving beyond purely scientific outcome predictions, but also to take into consideration what the patient's experience is, and to judge what we do or to predict how we can improve what we do and propose to patients, based on their feedback about the quality of their care. I think this is one of the objectives we should collectively aim for.
We thank Prof. Bernhard and Dr. Margue for their time and for sharing their experience. Visit the UroPredict page to learn more about this machine learning model on real-world data for the prediction of kidney cancer recurrence.
SOPHiA GENETICS products are for Research Use Only, not for use in diagnostic procedures unless otherwise specified.
Explore this infographic summary to learn more about the machine learning model developed by Margue et al. for the prediction of disease-free survival in patients undergoing surgery for renal cell carcinoma.
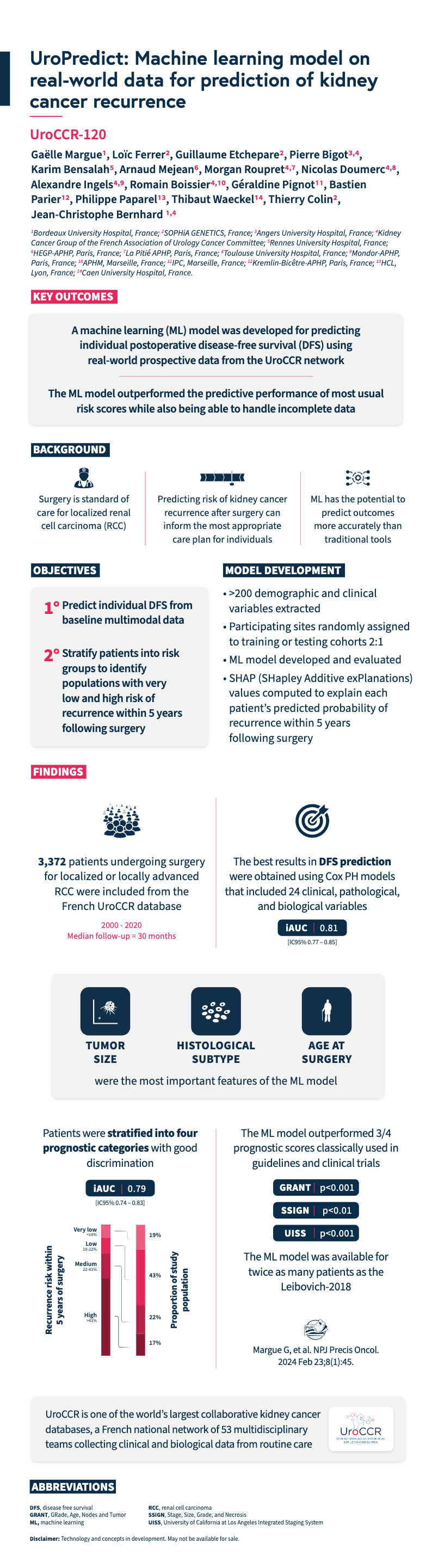
Margue G, et al. UroPredict: Machine learning model on real-world data for prediction of kidney cancer recurrence (UroCCR-120). NPJ Precis Oncol. 2024 Feb 23;8(1):45.
SOPHiA GENETICS multimodal analysis technology and concepts in development. May not be available for sale.
Do you want to further your understanding of overfitting, or are you interested to learn how we avoid overfitting when developing predictive machine learning models for clinical research? Browse our tech note, which includes a step-by-step, published example of how we developed a model that was able to successfully support the evaluation of kidney cancer tumor upstaging in individual patients.
Multimodal healthcare datasets synergistically integrate diverse data modalities such as genomic, clinical, radiomic, proteomic, and biological data, to provide comprehensive insights into human biology and medical conditions. Multimodal datasets have the potential to predict outcomes more accurately and informatively than the sum of their parts (Fig. 1).
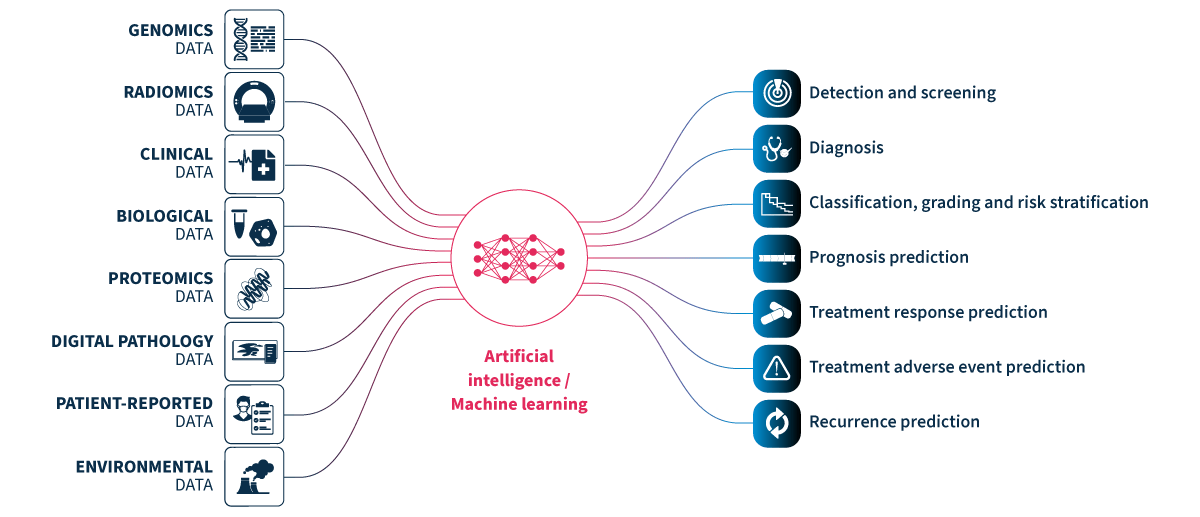
Figure 1.Multimodal healthcare data integrated and analyzed by artificial intelligence (AI)/machine learning can provide useful information for healthcare professionals to use to improve patient care.
Genomics data.
Radiomics data include x-rays, CT scans, MRI scans, ultrasound images, and mammograms.
Clinical and biological data from electronic health records include patient histories, demographics, notes, diagnosis codes, procedure codes, laboratory results, and vital signs.
Proteomics data.
Digital pathology data.
Patient-reported data includes questionnaires and health journals, as well as data from wearable devices monitoring heart rate, sleep patterns, and activity levels, and implantable devices such as pacemakers, insulin pumps, and continuous blood glucose monitors.
Environmental data includes air quality and location data.
New data-driven technologies powered by novel ways of linking and analyzing patient data are set to transform the way that healthcare is delivered.1 Healthcare professionals routinely make use of multiple sources of data to arrive at a diagnosis and to decide on patient management.2 However, a significant level of expertise is required for an in-depth understanding of even a single data type (e.g. radiological images) such that it is unfeasible for individual healthcare professionals to master all areas. AI/machine learning technologies can be leveraged to bring together and analyze multimodal healthcare data, breaking data silos and creating robust and accurate predictive models.3 With the appropriate guidance around decision-making and communication, the valuable insights gained from these predictive models have the potential to support healthcare professionals to improve patient care.
Machine learning technologies can integrate data from disparate multimodal sources to provide a holistic understanding of patients’ health and medical conditions. Data are combined from multiple modalities with the aim of extracting complementary information to power predictive models that can find relationships between different variables/features that are not clearly visible or known by healthcare professionals. Indeed, multimodal data fusion models have consistently shown to provide increased accuracy (1.2-27.7% higher) and performance (AUC 0.02-0.16 higher) than models that utilize data from single modalities for the same task.4
Oncology is one of the medical specialties that most commonly leverages multimodal methods for clinical decision support.5 Machine learning technologies have the potential to explore complex and diverse data to support healthcare professionals from screening to treatment (including relapse).6 Identification of risk factors can support non-invasive patient screening and preventive care.3 Detection of patterns in easily accessible data can help identify diagnostic or prognostic biomarkers to improve patient risk stratification or selection for clinical trials. Identification of predictive signatures of risk factors, adverse treatment reactions, treatment responses, or treatment benefit, can guide decisions around patient management.

Figure 2. The number of PubMed articles published on multimodal oncology data has dramatically increased in recent years.
PubMed search for ((multimodal) AND (oncology)) OR ((multimodal) AND (cancer)).
*2023 analysis includes data available at time of writing (January-September).
With data privacy and security paramount, multimodal healthcare data can also be leveraged to accelerate advances in medical research, such as the discovery of novel biomarkers and therapeutic targets for drug development, as well as supporting population health management by providing a comprehensive view of health trends and outcomes. The rapid increase in peer-reviewed publications on the topic over the last 13 years demonstrates that the extraordinary value of multimodal oncology data is already recognized by the scientific and medical communities (Fig. 2). Leveraging machine learning to collate and analyze the vast diversity of multimodal data for data-driven precision medicine is on track to drive the next revolution in healthcare.
SOPHiA DDM™ multimodal healthcare analytics will have the potential to break data silos by streamlining the integration of longitudinal oncology data from multiple sources and modalities – including but not limited to genomic, radiomic, digital pathology, biological, and clinical data. The SOPHiA DDM™ Platform uses machine learning-powered analytics to assemble, standardize, and transform multimodal data into accessible data-driven insights, facilitating the identification of multimodal predictive signatures, as well as treatment response patterns and trends. To learn more and get in touch, visit the webpage.
Product in development – Technology and concepts in development. May not be available for sale.
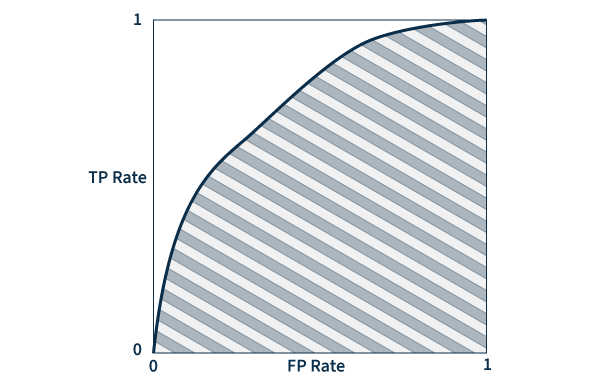
Area under the ROC curve (AUC) – A ROC (receiver operating characteristic) curve is a graph that plots true and false positive rates to demonstrate the performance of a model. AUC measures the area underneath the ROC curve to provide an aggregate measure of performance. AUC values range between 0 and 1, with a score of 0 meaning that all predictions are wrong, and a score of 1 meaning that all predictions are 100% correct. Essentially, AUC represents the probability that a positive result is truly positive and a negative result is truly negative.
Omics data – Large-scale information related to the biology of organisms.
Digital pathology images – Scanned images of tissue samples on glass slides.
Precision medicine, also known as personalized medicine, aims to enhance healthcare quality by tailoring treatments to each person's unique genetic makeup, environment, and lifestyle. While a fully individualized approach to medicine is still a work in progress, the recognition that patient heterogeneity influences treatment effectiveness is not new1.
Historically, medicine has heavily relied on trial-and-error strategies for discovering, developing, and testing new treatments targeted at specific indications. This disease-centered approach resulted in predetermined standard therapies tailored to the “average patient.” While this one-size-fits-all approach has succeeded in many indications, it also carries significant drawbacks, particularly when dealing with complex diseases such as cancer or inherited disorders. In these cases, the risk of adverse side effects (e.g., toxicity) and reduced therapeutic response often result in poorer patient prognoses and quality of life1.
Precision medicine represents a patient-centric paradigm shift, acknowledging each individual's uniqueness while using real-world data and advanced statistics to guide the discovery-to-treatment process. For instance, pharmacogenomics requires us to look at each patient genomic data individually and in the context of others, enabling stratification into cohorts for predicting treatment responses2.
Success in precision medicine hinges on the ability to derive meaningful insights from large patient datasets. Fostering data diversity has the potential to further advance progress in this area1,3.
Recent technological advances have made precision medicine more accessible and impactful than ever before. Next-generation sequencing (NGS) has become more affordable, transforming it from a research-focused technology into a tangible clinical reality. This progress was further propelled by the widespread adoption of electronic health records (EHRs) and laboratory information management systems (LIMS), which not only facilitate population-scale research but also enable the use of clinical decision support tools for the delivery of targeted therapies to individual patients4.
The ability to identify genetic biomarkers and assess variant pathogenicity has grown significantly in the past decade. This has not only revolutionized patient diagnosis but also transformed drug development. A pivotal moment was the approval of imatinib by the FDA in 2001, the first small molecule targeted therapy for chronic myeloid leukemia (CML)5. By inhibiting the BCR-ABL fusion protein, imatinib was shown to effectively slow the progression of CML from chronic phase to blast crisis, making it the first of its kind.
This groundbreaking milestone paved the way for the approval of many other targeted therapies, such as gefitinib targeting EGFR alterations associated with NSCLC (2003) and trastuzumab for HER2-positive breast cancer (2006). The pace of new targeted drug approvals continues to accelerate year after year, heralding a promising era of precision medicine6.
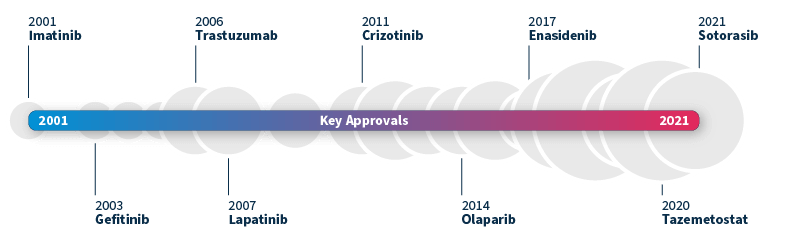
Timeline of FDA-approved targeted therapies in cancer. Grey bubbles represent the relative number of approvals per year. Data source: Waarts et al 2022.
To achieve a truly personalized approach to medicine, the harmonization of translational and precision medicine is paramount. This coordination between early mechanism-based drug development and late-stage patient-centric approaches gives rise to an end-to-end biomarker-guided process, allowing us to optimize treatment strategies for patient cohorts right from the outset7.
Known as translational precision medicine, this emerging concept brings a fresh perspective to the translational gap, calling for a broader scope beyond a purely genetic-based definition of biomarkers and introducing a multimodal approach by taking into account a wider range of healthcare variables. To make this new concept a reality, significant technological progress is required in several key areas8:
By addressing these critical areas of advancement, we can pave the way for a future where each patient receives personalized treatments tailored to her or his unique needs and characteristics. The pursuit of translational precision medicine promises to revolutionize healthcare, offering improved patient outcomes and transforming the landscape of medical research and development.
Powered by proprietary algorithms and enriched with data from 750+ institutions*, the SOPHiA DDM™ Platform accelerates advances in the field of precision medicine. Its core mission centers on empowering clinical researchers across healthcare and biopharma spheres alike.
To learn more about SOPHiA DDM™ BioPharma Solutions for biomarker-centric discovery, development, and application deployment, visit our page.
* The number of institutions represents active customers who have generated revenue through the SOPHiA DDM™ Platform usage or Alamut™ Visual Plus licenses as of September 30, 2022.
SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.